NAS API Reference¶
Model space¶
- class nni.nas.nn.pytorch.LayerChoice(*args, **kwargs)[source]¶
Layer choice selects one of the
candidates, then apply it on inputs and return results.It allows users to put several candidate operations (e.g., PyTorch modules), one of them is chosen in each explored model.
New in v2.2: Layer choice can be nested.
- Parameters:
candidates (list of nn.Module or OrderedDict) – A module list to be selected from.
weights (list of float) – Prior distribution used in random sampling.
label (str) – Identifier of the layer choice.
- length¶
Deprecated. Number of ops to choose from.
len(layer_choice)is recommended.- Type:
int
- names¶
Names of candidates.
- Type:
list of str
- choices¶
Deprecated. A list of all candidate modules in the layer choice module.
list(layer_choice)is recommended, which will serve the same purpose.- Type:
list of Module
Examples
# import nni.nas.nn.pytorch as nn # declared in `__init__` method self.layer = nn.LayerChoice([ ops.PoolBN('max', channels, 3, stride, 1), ops.SepConv(channels, channels, 3, stride, 1), nn.Identity() ]) # invoked in `forward` method out = self.layer(x)
Notes
candidatescan be a list of modules or a ordered dict of named modules, for example,self.op_choice = LayerChoice(OrderedDict([ ("conv3x3", nn.Conv2d(3, 16, 128)), ("conv5x5", nn.Conv2d(5, 16, 128)), ("conv7x7", nn.Conv2d(7, 16, 128)) ]))
Elements in layer choice can be modified or deleted. Use
del self.op_choice["conv5x5"]orself.op_choice[1] = nn.Conv3d(...). Adding more choices is not supported yet.- property candidates: Dict[str, Module] | List[Module]¶
Restore the
candidatesparameters passed to the constructor. Useful when creating a new layer choices based on this one.
- class nni.nas.nn.pytorch.InputChoice(*args, **kwargs)[source]¶
Input choice selects
n_choseninputs fromchoose_from(containsn_candidateskeys).It is mainly for choosing (or trying) different connections. It takes several tensors and chooses
n_chosentensors from them. When specific inputs are chosen,InputChoicewill becomeChosenInputs.Use
reductionto specify how chosen inputs are reduced into one output. A few options are:none: do nothing and return the list directly.sum: summing all the chosen inputs.mean: taking the average of all chosen inputs.concat: concatenate all chosen inputs at dimension 1.
We don’t support customizing reduction yet.
- Parameters:
n_candidates (int) – Number of inputs to choose from. It is required.
n_chosen (int) – Recommended inputs to choose. If None, mutator is instructed to select any.
reduction (str) –
mean,concat,sumornone.weights (list of float) – Prior distribution used in random sampling.
label (str) – Identifier of the input choice.
Examples
# import nni.nas.nn.pytorch as nn # declared in `__init__` method self.input_switch = nn.InputChoice(n_chosen=1) # invoked in `forward` method, choose one from the three out = self.input_switch([tensor1, tensor2, tensor3])
- class nni.nas.nn.pytorch.Repeat(*args, **kwargs)[source]¶
Repeat a block by a variable number of times.
- Parameters:
blocks (function, list of function, module or list of module) – The block to be repeated. If not a list, it will be replicated (deep-copied) into a list. If a list, it should be of length
max_depth, the modules will be instantiated in order and a prefix will be taken. If a function, it will be called (the argument is the index) to instantiate a module. Otherwise the module will be deep-copied.depth (int or tuple of int) –
If one number, the block will be repeated by a fixed number of times. If a tuple, it should be (min, max), meaning that the block will be repeated at least
mintimes and at mostmaxtimes. If a ValueChoice, it should choose from a series of positive integers.New in version 2.8: Minimum depth can be 0. But this feature is NOT supported on graph engine.
Examples
Block() will be deep copied and repeated 3 times.
self.blocks = nn.Repeat(Block(), 3)
Block() will be repeated 1, 2, or 3 times.
self.blocks = nn.Repeat(Block(), (1, 3))
Can be used together with layer choice. With deep copy, the 3 layers will have the same label, thus share the choice.
self.blocks = nn.Repeat(nn.LayerChoice([...]), (1, 3))
To make the three layer choices independent, we need a factory function that accepts index (0, 1, 2, …) and returns the module of the
index-th layer.self.blocks = nn.Repeat(lambda index: nn.LayerChoice([...], label=f'layer{index}'), (1, 3))
Depth can be a ValueChoice to support arbitrary depth candidate list.
self.blocks = nn.Repeat(Block(), nn.ValueChoice([1, 3, 5]))
- class nni.nas.nn.pytorch.Cell(*args, **kwargs)[source]¶
Cell structure that is popularly used in NAS literature.
Find the details in:
On Network Design Spaces for Visual Recognition is a good summary of how this structure works in practice.
A cell consists of multiple “nodes”. Each node is a sum of multiple operators. Each operator is chosen from
op_candidates, and takes one input from previous nodes and predecessors. Predecessor means the input of cell. The output of cell is the concatenation of some of the nodes in the cell (by default all the nodes).Two examples of searched cells are illustrated in the figure below. In these two cells,
op_candidatesare series of convolutions and pooling operations.num_nodes_per_nodeis set to 2.num_nodesis set to 5.merge_opisloose_end. Assuming nodes are enumerated from bottom to top, left to right,output_node_indicesfor the normal cell is[2, 3, 4, 5, 6]. For the reduction cell, it’s[4, 5, 6]. Please take a look at this review article if you are interested in details.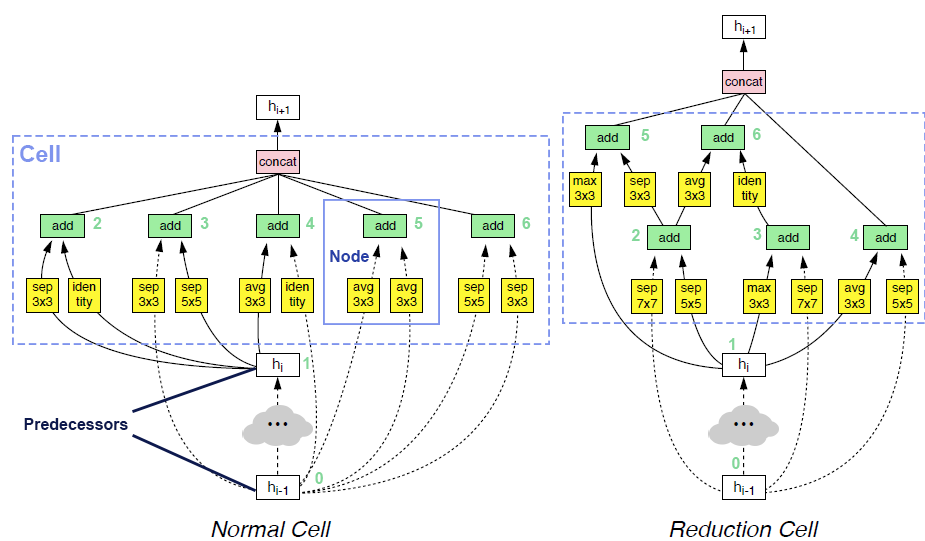
Here is a glossary table, which could help better understand the terms used above:
Name
Brief Description
Cell
A cell consists of
num_nodesnodes.Node
A node is the sum of
num_ops_per_nodeoperators.Operator
Each operator is independently chosen from a list of user-specified candidate operators.
Operator’s input
Each operator has one input, chosen from previous nodes as well as predecessors.
Predecessors
Input of cell. A cell can have multiple predecessors. Predecessors are sent to preprocessor for preprocessing.
Cell’s output
Output of cell. Usually concatenation of some nodes (possibly all nodes) in the cell. Cell’s output, along with predecessors, are sent to postprocessor for postprocessing.
Preprocessor
Extra preprocessing to predecessors. Usually used in shape alignment (e.g., predecessors have different shapes). By default, do nothing.
Postprocessor
Extra postprocessing for cell’s output. Usually used to chain cells with multiple Predecessors (e.g., the next cell wants to have the outputs of both this cell and previous cell as its input). By default, directly use this cell’s output.
Tip
It’s highly recommended to make the candidate operators have an output of the same shape as input. This is because, there can be dynamic connections within cell. If there’s shape change within operations, the input shape of the subsequent operation becomes unknown. In addition, the final concatenation could have shape mismatch issues.
- Parameters:
op_candidates (list of module or function, or dict) – A list of modules to choose from, or a function that accepts current index and optionally its input index, and returns a module. For example, (2, 3, 0) means the 3rd op in the 2nd node, accepts the 0th node as input. The index are enumerated for all nodes including predecessors from 0. When first created, the input index is
None, meaning unknown. Note that in graph execution engine, support of function inop_candidatesis limited. Please also note that, to makeCellwork with one-shot strategy,op_candidates, in case it’s a callable, should not depend on the second input argument, i.e.,op_indexin current node.num_nodes (int) – Number of nodes in the cell.
num_ops_per_node (int) – Number of operators in each node. The output of each node is the sum of all operators in the node. Default: 1.
num_predecessors (int) – Number of inputs of the cell. The input to forward should be a list of tensors. Default: 1.
merge_op ("all", or "loose_end") – If “all”, all the nodes (except predecessors) will be concatenated as the cell’s output, in which case,
output_node_indiceswill belist(range(num_predecessors, num_predecessors + num_nodes)). If “loose_end”, only the nodes that have never been used as other nodes’ inputs will be concatenated to the output. Predecessors are not considered when calculating unused nodes. Details can be found in NDS paper. Default: all.preprocessor (callable) – Override this if some extra transformation on cell’s input is intended. It should be a callable (
nn.Moduleis also acceptable) that takes a list of tensors which are predecessors, and outputs a list of tensors, with the same length as input. By default, it does nothing to the input.postprocessor (callable) – Override this if customization on the output of the cell is intended. It should be a callable that takes the output of this cell, and a list which are predecessors. Its return type should be either one tensor, or a tuple of tensors. The return value of postprocessor is the return value of the cell’s forward. By default, it returns only the output of the current cell.
concat_dim (int) – The result will be a concatenation of several nodes on this dim. Default: 1.
label (str) – Identifier of the cell. Cell sharing the same label will semantically share the same choice.
Examples
Choose between conv2d and maxpool2d. The cell have 4 nodes, 1 op per node, and 2 predecessors.
>>> cell = nn.Cell([nn.Conv2d(32, 32, 3, padding=1), nn.MaxPool2d(3, padding=1)], 4, 1, 2)
In forward:
>>> cell([input1, input2])
The “list bracket” can be omitted:
>>> cell(only_input) # only one input >>> cell(tensor1, tensor2, tensor3) # multiple inputs
Use
merge_opto specify how to construct the output. The output will then have dynamic shape, depending on which input has been used in the cell.>>> cell = nn.Cell([nn.Conv2d(32, 32, 3), nn.MaxPool2d(3)], 4, 1, 2, merge_op='loose_end') >>> cell_out_channels = len(cell.output_node_indices) * 32
The op candidates can be callable that accepts node index in cell, op index in node, and input index.
>>> cell = nn.Cell([ ... lambda node_index, op_index, input_index: nn.Conv2d(32, 32, 3, stride=2 if input_index < 1 else 1), ... ], 4, 1, 2)
Predecessor example:
class Preprocessor: def __init__(self): self.conv1 = nn.Conv2d(16, 32, 1) self.conv2 = nn.Conv2d(64, 32, 1) def forward(self, x): return [self.conv1(x[0]), self.conv2(x[1])] cell = nn.Cell([nn.Conv2d(32, 32, 3), nn.MaxPool2d(3)], 4, 1, 2, preprocessor=Preprocessor()) cell([torch.randn(1, 16, 48, 48), torch.randn(1, 64, 48, 48)]) # the two inputs will be sent to conv1 and conv2 respectively
Warning
Cellis not supported inGraphModelSpacemodel format.- output_node_indices¶
An attribute that contains indices of the nodes concatenated to the output (a list of integers).
When the cell is first instantiated in the base model, or when
merge_opisall,output_node_indicesmust berange(num_predecessors, num_predecessors + num_nodes).When
merge_opisloose_end,output_node_indicesis useful to compute the shape of this cell’s output, because the output shape depends on the connection in the cell, and which nodes are “loose ends” depends on mutation.- Type:
list of int
- op_candidates_factory¶
If the operations are created with a factory (callable), this is to be set with the factory. One-shot algorithms will use this to make each node a cartesian product of operations and inputs.
- Type:
CellOpFactory or None
- forward(*inputs)[source]¶
Forward propagation of cell.
- Parameters:
inputs (List[Tensor] | Tensor) – Can be a list of tensors, or several tensors. The length should be equal to
num_predecessors.- Returns:
The return type depends on the output of
postprocessor. By default, it’s the output ofmerge_op, which is a contenation (onconcat_dim) of some of (possibly all) the nodes’ outputs in the cell.- Return type:
Tuple[torch.Tensor] | torch.Tensor
- class nni.nas.nn.pytorch.ModelSpace(*args, **kwargs)[source]¶
The base class for model search space based on PyTorch. The out-est module should inherit this class.
Model space is written as PyTorch module for the convenience of writing code. It’s not a real PyTorch model, and shouldn’t be used as one for most cases. Most likely, the forward of
ModelSpaceis a dry run of an arbitrary model in the model space. But since there is no guarantee on which model will be chosen, and the behavior is not well tested, it’s only used for sanity check and tracing the space, and its semantics are not well-defined.Similarly for
state_dictandload_state_dict. Users should bear in mind thatModelSpaceis NOT a one-shot supernet, directly exporting its weights are unreliable and prone to error. Use one-shot strategies to mutate the model space into a supernet for such needs.Mutables in model space must all be labeled manually, unless a label prefix is provided. Every model space can have a label prefix, which is used to provide a stable automatic label generation. For example, if the label prefix is
model, all the mutables initialized in a subclass of ModelSpace (in__init__function of itself and submodules, to be specific), will be automatically labeled with a prefixmodel/. The label prefix can be manually specified upon definition of the class:class MyModelSpace(ModelSpace, label_prefix='backbone'): def __init__(self): super().__init__() self.choice = self.add_mutable(nni.choice('depth', [2, 3, 4])) print(self.choice.label) # backbone/choice
Notes
The
__init__implementation ofModelSpaceis inmodel_space_init_wrapper().
- class nni.nas.nn.pytorch.ParametrizedModule(*args, **kwargs)[source]¶
Subclass of
MutableModulesupports mutables as initialization parameters.One important feature of
ParametrizedModuleis that it automatically freeze the mutable arguments passed to__init__. This is for the convenience as well as compatibility with existing code:class MyModule(ParametrizedModule): def __init__(self, x): super().__init__() self.t = x # Will be a fixed number, e.g., 3. MyModule(nni.choice('choice1', [1, 2, 3]))
Note that the mutable arguments need to be directly posed as arguments to
__init__. They can’t be hidden in a list or dict.If users want to make a 3rd-party module parametrized, it’s recommended to do the following (taking
nn.Conv2das an example):>>> class ParametrizedConv2d(ParametrizedModule, nn.Conv2d, wraps=nn.Conv2d): ... pass >>> conv = ParametrizedConv2d(3, nni.choice('out', [8, 16])) >>> conv >>> conv.out_channels 8 >>> conv.args['out_channels'] Categorical([8, 16], label='out') >>> conv.freeze({'out': 16}) Conv2d(3, 16, kernel_size=(1, 1), stride=(1, 1))
Tip
The parametrized version of modules in
torch.nnare already provided innni.nas.nn.pytorch. Every class is prefixed withMutable. For example,nni.nas.nn.pytorch.MutableConv2d`is a parametrized version oftorch.nn.Conv2d.- args¶
The arguments used to initialize the module. Since
ParametrizedModulewill hijack the init arguments before passing to__init__, this is the only recommended way to retrieve the original init arguments back.
Warning
ParametrizedModulecan be nested. It’s also possible to put arbitrary mutable modules inside aParametrizedModule. But be careful if the inner mutable modules are dependant on the parameters ofParametrizedModule, because NNI can’t handle cases where the mutables are a dynamically changing after initialization. For example, the following snippet is WRONG:class MyModule(ParametrizedModule): def __init__(self, x): if x == 0: self.mutable = self.add_mutable(nni.choice('a', [1, 2, 3])) else: self.mutable = self.add_mutable(nni.choice('b', [4, 5, 6])) module = MyModule(nni.choice('x', [0, 1]))
- class nni.nas.nn.pytorch.MutableModule(*args, **kwargs)[source]¶
PyTorch module, but with uncertainties.
This base class provides useful tools to handle search spaces built on top of PyTorch modules, including methods like
simplify(),freeze().MutableModulecan have dangling mutables registered on it viaadd_mutable().- add_mutable(mutable)[source]¶
Register a mutable to this module. This is often used to add dangling variables that are not parameters of any
ParametrizedModule.If the mutable is also happens to be a submodule of type
MutableModule, it can be registered in the same way as PyTorch (i.e.,self.xxx = mutable). No need to add it again here.Examples
In practice, this method is often used together with
ensure_frozen().>>> class MyModule(MutableModule): ... def __init__(self): ... super().__init__() ... token_size = nni.choice('t', [4, 8, 16]) # Categorical variable here ... self.add_mutable(token_size) # Register the mutable to this module. ... real_token_size = ensure_frozen(token_size) # Real number. 4 during dry run. 4, 8 or 16 during search. ... self.token = nn.Parameter(torch.randn(real_token_size, 1))
Tip
Note that
ensure_frozen()must be used under afrozen_context(). The easiest way to do so is to invoke it within initialization of aModelSpace.Warning
Arbitrary
add_mutable()is not supported forGraphModelSpace.
- classmethod create_fixed_module(sample, *args, **kwargs)[source]¶
The classmethod is to create a brand new module with fixed architecture.
The parameter
sampleis a dict with the exactly same format assampleinfreeze(). The difference is that whencreate_fixed_module()is called, there is noMutableModuleinstance created yet. Thus it can be useful to simplify the creation of a fixed module, by saving the cost of creating aMutableModuleinstance and immediatelyfreeze()it.If automatic label generation (e.g.,
auto_label()) is used in__init__, the same number of labels should be generated in this method. Otherwise it will mess up the global label counter, and potentially affect the label of successive modules.By default, this method has a not-implemented flag, and
should_invoke_fixed_module()will returnFalsebased on this flag.
- freeze(sample)[source]¶
Return a frozen version of current mutable module. Some sub-modules can be possibly deep-copied.
If mutables are added to the module via
add_mutable(), this method must be implemented. Otherwise, it will simply look at the children modules and freeze them recursively.freeze()of subclass is encouraged to keep the original weights at best effort, but no guarantee is made, unless otherwise specified.
- mutable_descendants()[source]¶
named_mutable_descendants()without names.
- property mutables: List[Mutable]¶
Mutables that are dangling under this module.
Normally this is all the mutables that are registered via
MutableModule.add_mutable().
- named_mutable_descendants()[source]¶
Traverse the module subtree, find all descendants that are
MutableModule.If a child module is
MutableModule, return it directly, and its subtree will be ignored.If not, it will be recursively expanded, until
MutableModuleis found.
- classmethod should_invoke_fixed_module()[source]¶
Call
create_fixed_module()when fixed-arch context is detected.Typically this should be enabled. Otherwise the arch context might not be correctly handled. In cases where this flag is disabled, remember to detect arch context and manually freeze things in
__init__, or confirm that it’s a composite module and nothing needs to be frozen.By default, it returns true when
create_fixed_module()is overridden.
Model Space Hub¶
NasBench101¶
- class nni.nas.hub.pytorch.NasBench101(*args, **kwargs)[source]¶
The full search space proposed by NAS-Bench-101.
It’s simply a stack of
NasBench101Cell. Operations are conv3x3, conv1x1 and maxpool respectively.- Parameters:
stem_out_channels – Number of output channels of the stem convolution.
num_stacks – Number of stacks in the network.
num_modules_per_stack – Number of modules in each stack. Each module is a
NasBench101Cell.max_num_vertices – Maximum number of vertices in each cell.
max_num_edges – Maximum number of edges in each cell.
num_labels – Number of categories for classification.
bn_eps – Epsilon for batch normalization.
bn_momentum – Momentum for batch normalization.
NasBench201¶
- class nni.nas.hub.pytorch.NasBench201(*args, **kwargs)[source]¶
The full search space proposed by NAS-Bench-201.
It’s a stack of
NasBench201Cell.- Parameters:
stem_out_channels – The output channels of the stem.
num_modules_per_stack – The number of modules (cells) in each stack. Each cell is a
NasBench201Cell.num_labels – Number of categories for classification.
NASNet¶
- class nni.nas.hub.pytorch.NASNet(*args, **kwargs)[source]¶
Search space proposed in Learning Transferable Architectures for Scalable Image Recognition.
It is built upon
Cell, and implemented based onNDS. Its operator candidates areNASNET_OPS. It has 5 nodes per cell, and the output is concatenation of nodes not used as input to other nodes.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.nas.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width – A fixed initial width or a tuple of widths to choose from.
num_cells – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob – Apply drop path. Enabled when it’s set to be greater than 0.
- NASNET_OPS = ['skip_connect', 'conv_3x1_1x3', 'conv_7x1_1x7', 'dil_conv_3x3', 'avg_pool_3x3', 'max_pool_3x3', 'max_pool_5x5', 'max_pool_7x7', 'conv_1x1', 'conv_3x3', 'sep_conv_3x3', 'sep_conv_5x5', 'sep_conv_7x7']¶
The candidate operations.
- class nni.nas.hub.pytorch.nasnet.NDS(*args, **kwargs)[source]¶
The unified version of NASNet search space.
We follow the implementation in unnas. See On Network Design Spaces for Visual Recognition for details.
Different NAS papers usually differ in the way that they specify
op_candidatesandmerge_op.datasethere is to give a hint about input resolution, so as to create reasonable stem and auxiliary heads.NDS has a speciality that it has mutable depths/widths. This is implemented by accepting a list of int as
num_cells/width.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.nas.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width – A fixed initial width or a tuple of widths to choose from.
num_cells – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob – Apply drop path. Enabled when it’s set to be greater than 0.
op_candidates – List of operator candidates. Must be from
OPS.merge_op – See
Cell.num_nodes_per_cell – See
Cell.
- freeze(sample)[source]¶
Freeze the model according to the sample.
As different stages have dependencies among each other, we will recreate the whole model for simplicity. For weight inheritance purposes, this
freeze()might require re-writing.- Parameters:
sample (Dict[str, Any]) – The architecture dict.
- set_drop_path_prob(drop_prob)[source]¶
Set the drop probability of Drop-path in the network. Reference: FractalNet: Ultra-Deep Neural Networks without Residuals.
- class nni.nas.hub.pytorch.nasnet.NDSStage(*args, **kwargs)[source]¶
This class defines NDSStage, a special type of Repeat, for isinstance check, and shape alignment.
In NDS, we can’t simply use Repeat to stack the blocks, because the output shape of each stacked block can be different. This is a problem for one-shot strategy because they assume every possible candidate should return values of the same shape.
Therefore, we need
NDSStagePathSamplingandNDSStageDifferentiableto manually align the shapes – specifically, to transform the first block in each stage.This is not required though, when depth is not changing, or the mutable depth causes no problem (e.g., when the minimum depth is large enough).
Attention
Assumption: Loose end is treated as all in
merge_op(the case in one-shot), which enforces reduction cell and normal cells in the same stage to have the exact same output shape.- downsampling: bool¶
This stage has downsampling
- estimated_out_channels: int¶
Output channels of this stage. It’s estimated because it assumes
allasmerge_op.
- estimated_out_channels_prev: int¶
Output channels of cells in last stage.
ENAS¶
- class nni.nas.hub.pytorch.ENAS(*args, **kwargs)[source]¶
Search space proposed in Efficient neural architecture search via parameter sharing.
It is built upon
Cell, and implemented based onNDS. Its operator candidates areENAS_OPS. It has 5 nodes per cell, and the output is concatenation of nodes not used as input to other nodes.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.nas.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width – A fixed initial width or a tuple of widths to choose from.
num_cells – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob – Apply drop path. Enabled when it’s set to be greater than 0.
- ENAS_OPS = ['skip_connect', 'sep_conv_3x3', 'sep_conv_5x5', 'avg_pool_3x3', 'max_pool_3x3']¶
The candidate operations.
AmoebaNet¶
- class nni.nas.hub.pytorch.AmoebaNet(*args, **kwargs)[source]¶
Search space proposed in Regularized evolution for image classifier architecture search.
It is built upon
Cell, and implemented based onNDS. Its operator candidates areAMOEBA_OPS. It has 5 nodes per cell, and the output is concatenation of nodes not used as input to other nodes.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.nas.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width – A fixed initial width or a tuple of widths to choose from.
num_cells – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob – Apply drop path. Enabled when it’s set to be greater than 0.
- AMOEBA_OPS = ['skip_connect', 'sep_conv_3x3', 'sep_conv_5x5', 'sep_conv_7x7', 'avg_pool_3x3', 'max_pool_3x3', 'dil_sep_conv_3x3', 'conv_7x1_1x7']¶
The candidate operations.
PNAS¶
- class nni.nas.hub.pytorch.PNAS(*args, **kwargs)[source]¶
Search space proposed in Progressive neural architecture search.
It is built upon
Cell, and implemented based onNDS. Its operator candidates arePNAS_OPS. It has 5 nodes per cell, and the output is concatenation of all nodes in the cell.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.nas.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width – A fixed initial width or a tuple of widths to choose from.
num_cells – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob – Apply drop path. Enabled when it’s set to be greater than 0.
- PNAS_OPS = ['sep_conv_3x3', 'sep_conv_5x5', 'sep_conv_7x7', 'conv_7x1_1x7', 'skip_connect', 'avg_pool_3x3', 'max_pool_3x3', 'dil_conv_3x3']¶
The candidate operations.
DARTS¶
- class nni.nas.hub.pytorch.DARTS(*args, **kwargs)[source]¶
Search space proposed in Darts: Differentiable architecture search.
It is built upon
Cell, and implemented based onNDS. Its operator candidates areDARTS_OPS. It has 4 nodes per cell, and the output is concatenation of all nodes in the cell.Note
noneis not included in the operator candidates. It has already been handled in the differentiable implementation of cell.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.nas.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width – A fixed initial width or a tuple of widths to choose from.
num_cells – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob – Apply drop path. Enabled when it’s set to be greater than 0.
- DARTS_OPS = ['max_pool_3x3', 'avg_pool_3x3', 'skip_connect', 'sep_conv_3x3', 'sep_conv_5x5', 'dil_conv_3x3', 'dil_conv_5x5']¶
The candidate operations.
ProxylessNAS¶
- class nni.nas.hub.pytorch.ProxylessNAS(*args, **kwargs)[source]¶
The search space proposed by ProxylessNAS.
Following the official implementation, the inverted residual with kernel size / expand ratio variations in each layer is implemented with a
LayerChoicewith all-combination candidates. That means, when used in weight sharing, these candidates will be treated as separate layers, and won’t be fine-grained shared. We note thatMobileNetV3Spaceis different in this perspective.This space can be implemented as part of
MobileNetV3Space, but we separate those following conventions.- Parameters:
num_labels – The number of labels for classification.
base_widths – Widths of each stage, from stem, to body, to head. Length should be 9.
dropout_rate – Dropout rate for the final classification layer.
width_mult – Width multiplier for the model.
bn_eps – Epsilon for batch normalization.
bn_momentum – Momentum for batch normalization.
- class nni.nas.hub.pytorch.proxylessnas.InvertedResidual(in_channels, out_channels, expand_ratio, kernel_size=3, stride=1, squeeze_excite=None, norm_layer=None, activation_layer=None)[source]¶
An Inverted Residual Block, sometimes called an MBConv Block, is a type of residual block used for image models that uses an inverted structure for efficiency reasons.
It was originally proposed for the MobileNetV2 CNN architecture. It has since been reused for several mobile-optimized CNNs. It follows a narrow -> wide -> narrow approach, hence the inversion. It first widens with a 1x1 convolution, then uses a 3x3 depthwise convolution (which greatly reduces the number of parameters), then a 1x1 convolution is used to reduce the number of channels so input and output can be added.
This implementation is sort of a mixture between:
- Parameters:
in_channels (int | MutableExpression[int]) – The number of input channels. Can be a value choice.
out_channels (int | MutableExpression[int]) – The number of output channels. Can be a value choice.
expand_ratio (float | MutableExpression[float]) – The ratio of intermediate channels with respect to input channels. Can be a value choice.
kernel_size (int | MutableExpression[int]) – The kernel size of the depthwise convolution. Can be a value choice.
stride (int) – The stride of the depthwise convolution.
squeeze_excite (Callable[[int | MutableExpression[int], int | MutableExpression[int]], Module] | None) – Callable to create squeeze and excitation layer. Take hidden channels and input channels as arguments.
norm_layer (Callable[[int], Module] | None) – Callable to create normalization layer. Take input channels as argument.
activation_layer (Callable[[...], Module] | None) – Callable to create activation layer. No input arguments.
MobileNetV3Space¶
- class nni.nas.hub.pytorch.MobileNetV3Space(*args, **kwargs)[source]¶
MobileNetV3Space implements the largest search space in TuNAS.
The search dimensions include widths, expand ratios, kernel sizes, SE ratio. Some of them can be turned off via arguments to narrow down the search space.
Different from ProxylessNAS search space, this space is implemented with
ValueChoice.We use the following snipppet as reference. https://github.com/google-research/google-research/blob/20736344591f774f4b1570af64624ed1e18d2867/tunas/mobile_search_space_v3.py#L728
We have
num_blockswhich equals to the length ofself.blocks(the main body of the network). For simplicity, the following parameter specification assumesnum_blocksequals 8 (body + head). If a shallower body is intended, arrays includingbase_widths,squeeze_excite,depth_range,stride,activationshould also be shortened accordingly.- Parameters:
num_labels – Dimensions for classification head.
base_widths – Widths of each stage, from stem, to body, to head. Length should be 9, i.e.,
num_blocks + 1(because there is a stem width in front).width_multipliers – A range of widths multiplier to choose from. The choice is independent for each stage. Or it can be a fixed float. This will be applied on
base_widths, and we would also make sure that widths can be divided by 8.expand_ratios – A list of expand ratios to choose from. Independent for every block.
squeeze_excite – Indicating whether the current stage can have an optional SE layer. Expect array of length 6 for stage 0 to 5. Each element can be one of
force,optional,none.depth_range (List[Tuple[int, int]]) – A range (e.g.,
(1, 4)), or a list of range (e.g.,[(1, 3), (1, 4), (1, 4), (1, 3), (0, 2)]). If a list, the length should be 5. The depth are specified for stage 1 to 5.stride – Stride for all stages (including stem and head). Length should be same as
base_widths.activation – Activation (class) for all stages. Length is same as
base_widths.se_from_exp – Calculate SE channel reduction from expanded (mid) channels.
dropout_rate – Dropout rate at classification head.
bn_eps – Epsilon of batch normalization.
bn_momentum – Momentum of batch normalization.
ShuffleNetSpace¶
- class nni.nas.hub.pytorch.ShuffleNetSpace(*args, **kwargs)[source]¶
The search space proposed in Single Path One-shot.
The basic building block design is inspired by a state-of-the-art manually-designed network – ShuffleNetV2. There are 20 choice blocks in total. Each choice block has 4 candidates, namely
choice 3,choice 5,choice_7andchoice_xrespectively. They differ in kernel sizes and the number of depthwise convolutions. The size of the search space is \(4^{20}\).- Parameters:
num_labels (int) – Number of classes for the classification head. Default: 1000.
channel_search (bool) – If true, for each building block, the number of
mid_channels(output channels of the first 1x1 conv in each building block) varies from 0.2x to 1.6x (quantized to multiple of 0.2). Here, “k-x” means k times the number of default channels. Otherwise, 1.0x is used by default. Default: false.affine (bool) – Apply affine to all batch norm. Default: true.
AutoFormer¶
- class nni.nas.hub.pytorch.AutoFormer(*args, **kwargs)[source]¶
The search space that is proposed in AutoFormer. There are four searchable variables: depth, embedding dimension, heads number and MLP ratio.
- Parameters:
search_embed_dim – The search space of embedding dimension. Use a list to specify search range.
search_mlp_ratio – The search space of MLP ratio. Use a list to specify search range.
search_num_heads – The search space of number of heads. Use a list to specify search range.
search_depth – The search space of depth. Use a list to specify search range.
img_size – Size of input image.
patch_size – Size of image patch.
in_channels – Number of channels of the input image.
num_labels – Number of classes for classifier.
qkv_bias – Whether to use bias item in the qkv embedding.
drop_rate – Drop rate of the MLP projection in MSA and FFN.
attn_drop_rate – Drop rate of attention.
drop_path_rate – Drop path rate.
pre_norm – Whether to use pre_norm. Otherwise post_norm is used.
global_pooling – Whether to use global pooling to generate the image representation. Otherwise the cls_token is used.
absolute_position – Whether to use absolute positional embeddings.
qk_scale – The scaler on score map in self-attention.
rpe – Whether to use relative position encoding.
- classmethod load_pretrained_supernet(name, download=True, progress=True)[source]¶
Load the related supernet checkpoints.
Thanks to the weight entangling strategy that AutoFormer uses, AutoFormer releases a few trained supernet that allows thousands of subnets to be very well-trained. Under different constraints, different subnets can be found directly from the supernet, and used without any fine-tuning.
- Parameters:
name (str) – Search space size, must be one of {‘random-one-shot-tiny’, ‘random-one-shot-small’, ‘random-one-shot-base’}.
download (bool) – Whether to download supernet weights.
progress (bool) – Whether to display the download progress.
- Return type:
The loaded supernet.
- classmethod load_searched_model(name, pretrained=False, download=True, progress=True)[source]¶
Load the searched subnet model.
- Parameters:
name (str) – Search space size, must be one of {‘autoformer-tiny’, ‘autoformer-small’, ‘autoformer-base’}.
pretrained (bool) – Whether initialized with pre-trained weights.
download (bool) – Whether to download supernet weights.
progress (bool) – Whether to display the download progress.
- Returns:
The subnet model.
- Return type:
nn.Module
Module Components¶
Famous building blocks of search spaces.
- class nni.nas.hub.pytorch.modules.AutoActivation(*args, **kwargs)[source]¶
This module is an implementation of the paper Searching for Activation Functions.
- Parameters:
unit_num (int) – The number of core units.
unary_candidates (list[str] | None) – Names of unary candidates. If none, all names from
available_unary_choices()will be used.binary_candidates (list[str] | None) – Names of binary candidates. If none, all names from
available_binary_choices()will be used.label (str | None) – Label of the current module.
Notes
Currently,
beta(in operators likeBinaryParamAdd) is not per-channel parameter.
- class nni.nas.hub.pytorch.modules.NasBench101Cell(*args, **kwargs)[source]¶
Cell structure that is proposed in NAS-Bench-101.
Proposed by NAS-Bench-101: Towards Reproducible Neural Architecture Search.
This cell is usually used in evaluation of NAS algorithms because there is a “comprehensive analysis” of this search space available, which includes a full architecture-dataset that “maps 423k unique architectures to metrics including run time and accuracy”. You can also use the space in your own space design, in which scenario it should be possible to leverage results in the benchmark to narrow the huge space down to a few efficient architectures.
The space of this cell architecture consists of all possible directed acyclic graphs on no more than
max_num_nodesnodes, where each possible node (other than IN and OUT) has one ofop_candidates, representing the corresponding operation. Edges connecting the nodes can be no more thanmax_num_edges. To align with the paper settings, two vertices specially labeled as operation IN and OUT, are also counted intomax_num_nodesin our implementation, the default value ofmax_num_nodesis 7 andmax_num_edgesis 9.Input of this cell should be of shape \([N, C_{in}, *]\), while output should be \([N, C_{out}, *]\). The shape of each hidden nodes will be first automatically computed, depending on the cell structure. Each of the
op_candidatesshould be a callable that accepts computednum_featuresand returns aModule. For example,def conv_bn_relu(num_features): return nn.Sequential( nn.Conv2d(num_features, num_features, 1), nn.BatchNorm2d(num_features), nn.ReLU() )
The output of each node is the sum of its input node feed into its operation, except for the last node (output node), which is the concatenation of its input hidden nodes, adding the IN node (if IN and OUT are connected).
When input tensor is added with any other tensor, there could be shape mismatch. Therefore, a projection transformation is needed to transform the input tensor. In paper, this is simply a Conv1x1 followed by BN and ReLU. The
projectionparameters acceptsin_featuresandout_features, returns aModule. This parameter has no default value, as we hold no assumption that users are dealing with images. An example for this parameter is,def projection_fn(in_features, out_features): return nn.Conv2d(in_features, out_features, 1)
- Parameters:
op_candidates (list of callable) – Operation candidates. Each should be a function accepts number of feature, returning nn.Module.
in_features (int) – Input dimension of cell.
out_features (int) – Output dimension of cell.
projection (callable) – Projection module that is used to preprocess the input tensor of the whole cell. A callable that accept input feature and output feature, returning nn.Module.
max_num_nodes (int) – Maximum number of nodes in the cell, input and output included. At least 2. Default: 7.
max_num_edges (int) – Maximum number of edges in the cell. Default: 9.
label (str) – Identifier of the cell. Cell sharing the same label will semantically share the same choice.
Warning
NasBench101Cellis not supported for graph-based model format. It’s also not supported by most one-shot algorithms currently.
- class nni.nas.hub.pytorch.modules.NasBench201Cell(*args, **kwargs)[source]¶
Cell structure that is proposed in NAS-Bench-201.
Proposed by NAS-Bench-201: Extending the Scope of Reproducible Neural Architecture Search.
This cell is a densely connected DAG with
num_tensorsnodes, where each node is tensor. For every \(i < j\), there is an edge from i-th node to j-th node. Each edge in this DAG is associated with an operation transforming the hidden state from the source node to the target node. All possible operations are selected from a predefined operation set, defined inop_candidates. Each of theop_candidatesshould be a callable that accepts input dimension and output dimension, and returns aModule.Input of this cell should be of shape \([N, C_{in}, *]\), while output should be \([N, C_{out}, *]\). For example,
The space size of this cell would be \(|op|^{N(N-1)/2}\), where \(|op|\) is the number of operation candidates, and \(N\) is defined by
num_tensors.- Parameters:
op_candidates (list of callable) – Operation candidates. Each should be a function accepts input feature and output feature, returning nn.Module.
in_features (int) – Input dimension of cell.
out_features (int) – Output dimension of cell.
num_tensors (int) – Number of tensors in the cell (input included). Default: 4
label (str) – Identifier of the cell. Cell sharing the same label will semantically share the same choice.
Evaluator¶
- class nni.nas.evaluator.FunctionalEvaluator(function, **kwargs)[source]¶
Functional evaluator that directly takes a function and thus should be general. See
Evaluatorfor instructions on how to write this function.- function¶
The full name of the function.
- arguments¶
Keyword arguments for the function other than model.
- class nni.nas.evaluator.Evaluator[source]¶
Base class of evaluator.
To users, the evaluator is to assess the quality of a model and return a score. When an evaluator is defined, it usually accepts a few arguments, such as basic runtime information (e.g., whether to use GPU), dataset used, as well as hyper-parameters (such as learning rate). These parameters can be sometimes tunable and searched by algorithms (see
MutableEvaluator).Different evaluators could have different use scenarios and requirements on the model. For example,
Classificationis tailored for classification models, and assumes the model has aforwardmethod that takes a batch of data and returns logits. Evaluators might also have different assumptions, some of which are requirements of certain algorithms. The evaluator with the most freedom isFunctionalEvaluator, but it’s also incompatible with some algorithms.To developers, the evaluator is to implement all the logics involving forward/backward of neural networks. Sometimes the algorithm requires the training and searching at the same time (e.g., one-shot algos). In that case, although the searching part doesn’t logically belong to the evaluator, it is still the evaluator’s responsibility to implement it, and the search algorithms will make sure to properly manipulate the evaluator to achieve the goal.
Tip
Inside evaluator, you can use standard NNI trial APIs to communicate with the exploration strategy. Common usages include:
Use
nni.get_current_parameter()to get the currentExecutableModelSpace. Notice thatExecutableModelSpaceis not a directly-runnable model (e.g., a PyTorch model), which is different from the model received inevaluate().ExecutableModelSpaceobjects are useful for debugging, as well as for some evaluators which need to know extra details about how the model is sampled.Use
nni.report_intermediate_result()to report intermediate results.Use
nni.report_final_result()to report final results.
These APIs are only available when the evaluator is executed by NNI. We recommend using
nni.get_current_parameter() is not Noneto check if the APIs are available before using them. Please AVOID usingnni.get_next_parameter()because NAS framework has already handled the logic of retrieving the next parameter. Incorrectly usingnni.get_next_parameter()may cause unexpected behavior.- evaluate(model)[source]¶
To run evaluation of a model. The model is usually a concrete model. The return value of
evaluate()can be anything. Typically it’s used for test purposes.Subclass should override this.
- static mock_runtime(model)[source]¶
Context manager to mock trial APIs for standalone usage.
Under the with-context of this method,
nni.get_current_parameter()will return the given model.NOTE: This method might become a utility in trial command channel in future.
- Parameters:
model (ExecutableModelSpace) – The model to be evaluated. It should be a
ExecutableModelSpaceobject.
Examples
This method should be mostly used when testing a evaluator. A typical use case is as follows:
>>> frozen_model = model_space.freeze(sample) >>> with evaluator.mock_runtime(frozen_model): ... evaluator.evaluate(frozen_model.executable_model())
- class nni.nas.evaluator.MutableEvaluator[source]¶
Evaluators with tunable parameters by itself (e.g., learning rate).
The tunable parameters must be an argument of the evaluator’s instantiation, or an argument of the arguments’ instantiation and etc.
To use this feature, there are two requirements:
The evaluator must inherit
MutableEvaluatorrather thanEvaluator.Make sure the init arguments have been saved in
trace_kwargs, and the instance can be cloned withtrace_copy. The easiest way is to wrap the evaluator withnni.trace(). If the mutable parameter exists somewhere in the nested instantiation. All the levels must all be wrapped withnni.trace().
Examples
>>> def get_data(shuffle): ... ... >>> @nni.trace # 1. must wrap here ... class MyOwnEvaluator(MutableEvaluator): # 2. must inherit MutableEvaluator ... def __init__(self, lr, data): ... ... >>> evaluator = MyOwnEvaluator( ... lr=Categorical([0.1, 0.01]), # the argument can be tunable ... data=nni.trace(get_data)( # if there is mutable parameters inside, this must also have nni.trace ... shuffle=Categorical([False, True]) ... ) ... ) >>> evaluator.simplify() {'global/1': Categorical([0.1, 0.01], label='global/1'), 'global/2': Categorical([False, True], label='global/2')}
- freeze(sample)[source]¶
Upon freeze,
MutableEvaluatorwill freeze all the mutable parameters (as well as nested parameters), and return aFrozenEvaluator.The evaluator will not be fully initialized to save the memory, especially when parameters contain large objects such as datasets. To use the evaluator, call
FrozenEvaluator.get()to get the full usable evaluator.- Return type:
The frozen evaluator.
- class nni.nas.evaluator.pytorch.Classification(*args, **kwargs)[source]¶
Evaluator that is used for classification.
Available callback metrics in
Classificationare:train_loss
train_acc
val_loss
val_acc
- Parameters:
criterion (nn.Module) – Class for criterion module (not an instance). default:
nn.CrossEntropyLosslearning_rate (float) – Learning rate. default: 0.001
weight_decay (float) – L2 weight decay. default: 0
optimizer (Optimizer) – Class for optimizer (not an instance). default:
Adamtrain_dataloaders (DataLoader) – Used in
trainer.fit(). A PyTorch DataLoader with training samples. If thelightning_modulehas a predefined train_dataloader method this will be skipped.val_dataloaders (DataLoader or List of DataLoader) – Used in
trainer.fit(). Either a single PyTorch Dataloader or a list of them, specifying validation samples. If thelightning_modulehas a predefined val_dataloaders method this will be skipped.datamodule (LightningDataModule | None) – Used in
trainer.fit(). See Lightning DataModule.export_onnx (bool) – If true, model will be exported to
model.onnxbefore training starts. default truenum_classes (int) – Number of classes for classification task. Required for torchmetrics >= 0.11.0. default: None
trainer_kwargs (dict) – Optional keyword arguments passed to trainer. See Lightning documentation for details.
Examples
>>> evaluator = Classification()
To use customized criterion and optimizer:
>>> evaluator = Classification(nn.LabelSmoothingCrossEntropy, optimizer=torch.optim.SGD)
Extra keyword arguments will be passed to trainer, some of which might be necessary to enable GPU acceleration:
>>> evaluator = Classification(accelerator='gpu', devices=2, strategy='ddp')
- class nni.nas.evaluator.pytorch.Regression(*args, **kwargs)[source]¶
Evaluator that is used for regression.
Available callback metrics in
Regressionare:train_loss
train_mse
val_loss
val_mse
- Parameters:
criterion (nn.Module) – Class for criterion module (not an instance). default:
nn.MSELosslearning_rate (float) – Learning rate. default: 0.001
weight_decay (float) – L2 weight decay. default: 0
optimizer (Optimizer) – Class for optimizer (not an instance). default:
Adamtrain_dataloaders (DataLoader) – Used in
trainer.fit(). A PyTorch DataLoader with training samples. If thelightning_modulehas a predefined train_dataloader method this will be skipped.val_dataloaders (DataLoader or List of DataLoader) – Used in
trainer.fit(). Either a single PyTorch Dataloader or a list of them, specifying validation samples. If thelightning_modulehas a predefined val_dataloaders method this will be skipped.datamodule (LightningDataModule | None) – Used in
trainer.fit(). See Lightning DataModule.export_onnx (bool) – If true, model will be exported to
model.onnxbefore training starts. default: truetrainer_kwargs (dict) – Optional keyword arguments passed to trainer. See Lightning documentation for details.
Examples
>>> evaluator = Regression()
Extra keyword arguments will be passed to trainer, some of which might be necessary to enable GPU acceleration:
>>> evaluator = Regression(gpus=1)
- class nni.nas.evaluator.pytorch.Trainer(*args, **kwargs)[source]¶
Traced version of
pytorch_lightning.Trainer. See https://pytorch-lightning.readthedocs.io/en/stable/common/trainer.html
- class nni.nas.evaluator.pytorch.DataLoader(*args, **kwargs)[source]¶
Traced version of
torch.utils.data.DataLoader. See https://pytorch.org/docs/stable/data.html
- class nni.nas.evaluator.pytorch.Lightning(*args, **kwargs)[source]¶
Delegate the whole training to PyTorch Lightning.
Since the arguments passed to the initialization needs to be serialized,
LightningModule,TrainerorDataLoaderin this file should be used. Another option is to hide dataloader in the Lightning module, in which case, dataloaders are not required for this class to work.Following the programming style of Lightning, metrics sent to NNI should be obtained from
callback_metricsin trainer. Two hooks are added at the end of validation epoch and the end offit, respectively. The metric name and type depend on the specific task.Warning
The Lightning evaluator are stateful. If you try to use a previous Lightning evaluator, please note that the inner
lightning_moduleandtrainerwill be reused.- Parameters:
lightning_module (LightningModule) – Lightning module that defines the training logic.
trainer (Trainer) – Lightning trainer that handles the training.
train_dataloders – Used in
trainer.fit(). A PyTorch DataLoader with training samples. If thelightning_modulehas a predefined train_dataloader method this will be skipped. It can be any types of dataloader supported by Lightning.val_dataloaders (Any | None) – Used in
trainer.fit(). Either a single PyTorch Dataloader or a list of them, specifying validation samples. If thelightning_modulehas a predefined val_dataloaders method this will be skipped. It can be any types of dataloader supported by Lightning.datamodule (LightningDataModule | None) – Used in
trainer.fit(). See Lightning DataModule.fit_kwargs (Dict[str, Any] | None) – Keyword arguments passed to
trainer.fit().detect_interrupt (bool) – Lightning has a graceful shutdown mechanism. It does not terminate the whole program (but only the training) when a KeyboardInterrupt is received. Setting this to
Truewill raise the KeyboardInterrupt to the main process, so that the whole program can be terminated.
Examples
Users should define a Lightning module that inherits
LightningModule, and useTrainerandDataLoaderfrom`nni.nas.evaluator.pytorch, and make them parameters of this evaluator:import nni from nni.nas.evaluator.pytorch.lightning import Lightning, LightningModule, Trainer, DataLoader
- class nni.nas.evaluator.pytorch.LightningModule(*args, **kwargs)[source]¶
Basic wrapper of generated model. Lightning modules used in NNI should inherit this class.
It’s a subclass of
pytorch_lightning.LightningModule. See https://pytorch-lightning.readthedocs.io/en/stable/common/lightning_module.htmlSee
SupervisedLearningModuleas an example.- property model: Module¶
The inner model (architecture) to train / evaluate.
It will be only available after calling
set_model().
Multi-trial strategy¶
- class nni.nas.strategy.GridSearch(*, shuffle=True, seed=None, dedup=True)[source]¶
Traverse the search space and try all the possible combinations one by one.
- Parameters:
shuffle (bool) – Shuffle the order in a candidate list, so that they are tried in a random order. Currently, the implementation is a pseudo-random shuffle, which only shuffles the order of every 100 candidates.
seed (int | None) – Random seed.
- class nni.nas.strategy.Random(*, dedup=True, seed=None, **kwargs)[source]¶
Random search on the search space.
- Parameters:
dedup (bool) – Do not try the same configuration twice.
seed (int | None) – Random seed.
- class nni.nas.strategy.RegularizedEvolution(*, population_size=100, sample_size=25, mutation_prob=0.05, crossover=False, dedup=True, seed=None, **kwargs)[source]¶
Algorithm for regularized evolution (i.e. aging evolution). Follows “Algorithm 1” in Real et al. “Regularized Evolution for Image Classifier Architecture Search”, with several enhancements.
Sample in this algorithm are called individuals. Specifically, the first
population_sizeindividuals are randomly sampled from the search space, and the rest are generated via a selection and mutation process. While new individuals are added to the population, the oldest one is removed to keep the population size constant.- Parameters:
population_size (int) – The number of individuals to keep in the population.
sample_size (int) – The number of individuals that should participate in each tournament. When mutate,
sample_sizeindividuals can randomly selected from the population, and the best one among them will be treated as the parent.mutation_prob (float) – Probability that mutation happens in each dim.
crossover (bool) – If
True, the new individual will be a crossover between winners of two individual tournament. That means, two sets ofsample_sizeindividuals will be randomly selected from the population, and the best one in each set will be used as parents. Every dimension will be randomly selected from one of the parents.dedup (bool) – Enforce one sample to never appear twice. The population might be smaller than
population_sizeif this is set toTrueand the search space is small.seed (int | None) – Random seed.
- class nni.nas.strategy.PolicyBasedRL(*, samples_per_update=20, replay_buffer_size=None, reward_for_invalid=None, policy_fn=None, update_kwargs=None, **kwargs)[source]¶
Algorithm for policy-based reinforcement learning. This is a wrapper of algorithms provided in tianshou (PPO by default), and can be easily customized with other algorithms that inherit
BasePolicy(e.g., REINFORCE as in this paper).- Parameters:
samples_per_update (int) – How many models (trajectories) each time collector collects. After each collect, trainer will sample batch from replay buffer and do the update.
replay_buffer_size (int | None) – Size of replay buffer. If it’s none, the size will be the expected trajectory length times
samples_per_update.reward_for_invalid (float | None) – The reward for a sample that didn’t pass validation, or the training doesn’t return a metric. If not provided, failed models will be simply ignored as if nothing happened.
policy_fn (Optional[PolicyFactory]) – Since environment is created on the fly, the policy needs to be a factory function that creates a policy on-the-fly. It takes
TuningEnvironmentas input and returns a policy. By default, it will use the policy returned bydefault_policy_fn().update_kwargs (dict | None) – Keyword arguments for
policy.update. See tianshou’s BasePolicy for details. There is a special key"update_times"that can be used to specify how many timespolicy.updateis called, which can be used to sufficiently exploit the current available trajectories in the replay buffer (for example when actor and critic needs to be updated alternatively multiple times). By default, it’s{'batch_size': 32, 'repeat': 5, 'update_times': 5}.
- class nni.nas.strategy.TPE(*args, **kwargs)[source]¶
The Tree-structured Parzen Estimator (TPE) is a sequential model-based optimization (SMBO) approach.
Find the details in Algorithms for Hyper-Parameter Optimization.
SMBO methods sequentially construct models to approximate the performance of hyperparameters based on historical measurements, and then subsequently choose new hyperparameters to test based on this model.
Advanced APIs¶
Base¶
- class nni.nas.strategy.base.Strategy(model_space=None, engine=None)[source]¶
Base class for NAS strategies.
To explore a space with a strategy, use:
strategy = MyStrategy() strategy(model_space, engine)
The strategy has a
run()method, that defines the process of exploring a NAS space.Strategy is stateful. It might store information of the current
initialize()andrun()as member attributes. We do not allowrun()a strategy twice with same, or different model spaces.Subclass should override
_initialize()and_run(), as well asstate_dict()andload_state_dict()for checkpointing.- property engine: ExecutionEngine¶
Strategy should use
engineto submit models, listen to metrics, and do budget / concurrency control.The engine is set by
set_engine(), either manually, or by a NAS experiment.The engine could be either a real engine, or a middleware that wraps a real engine. It doesn’t make any difference because their interface are the same.
See also
- initialize(model_space, engine)[source]¶
Initialize the strategy.
This method should be called before
run()to initialize some states.Some strategies might even mutate the
model_space. They should return the mutated model space.load_state_dict()can be called afterinitialize()to restore the state of the strategy.Subclass override
_initialize()instead of this method.
- list_models(sort=True, limit=None)[source]¶
List all the models that is ever searched by the engine.
A typical use case of this is to get the top-performing models produced during
run().The default implementation uses
list_models()to retrieve a list of models from the execution engine.- Parameters:
sort (bool) – Whether to sort the models by their metric (in descending order). If sorted is true, only models with “Trained” status and non-
Nonemetric are returned.limit (int | None) – Limit the number of models to return.
- Return type:
An iterator of models.
- load_state_dict(state_dict)[source]¶
Load the state of the strategy. This is used for loading checkpoints.
The state of strategy is some variables that are related to the current exploration process. The loading is often done after
initialize()and beforerun().
- property model_space: ExecutableModelSpace¶
The model space that strategy is currently exploring.
It should be the same one as the input argument of
run(), but the property exists for convenience.See also
- run()[source]¶
Explore the model space.
This should be the main part of a NAS experiment. Strategies decide how to explore the model space. They can submit models to
enginefor training and evaluation.The strategy doesn’t have to wait for all the models it submits to finish training.
The caller of
run()is responsible of setting theengineandmodel_spacebefore callingrun().Subclass override
_run()instead of this method.
- class nni.nas.strategy.base.StrategyStatus(value)[source]¶
Status of a strategy.
A strategy is in one of the following statuses:
EMPTY: The strategy is not initialized.INITIALIZED: The strategy is initialized (with a model space), but not started.RUNNING: The strategy is running.SUCCEEDED: The strategy has successfully ended.INTERRUPTED: The strategy is interrupted.FAILED: The strategy is stopped due to error.
Middleware¶
- class nni.nas.strategy.middleware.Chain(strategy, *middlewares)[source]¶
Chain a
Strategy(main strategy) with severalStrategyMiddleware.All the communications between strategy and execution engine will pass through the chain of middlewares. For example, when the strategy submits a model, it will be handled by the middleware, which decides whether to hand over to the next middleware, or to manipulate, or even block the model. The last middleware is connected to the real execution engine (which might be also guarded by a few middlewares).
- Parameters:
strategy (Strategy) – The main strategy. There can be exactly one strategy which is submitting models actively, which is therefore called main strategy.
*middlewares (StrategyMiddleware) – A chain of middlewares. At least one.
See also
- class nni.nas.strategy.middleware.Deduplication(action, patience=1000, retain_history=True)[source]¶
This middleware is able to deduplicate models that are submitted by strategies.
When duplicated models are found, the middleware can be configured to, either mark the model as invalid, or find the metric of the model from history and “replay” the metrics. Regardless of which action is taken, the patience counter will always increase, and when it runs out, the middleware will say there is no more budget.
Notice that some strategies have already provided deduplication on their own, e.g.,
Random. This class is to help those strategies who do NOT have the ability of deduplication.- Parameters:
action (Literal['invalid', 'replay']) – What to do when a duplicated model is found.
invalidmeans to mark the model as invalid, whilereplaymeans to retrieve the metric of the previous same model from the engine.patience (int) – Number of continuous duplicated models received until the middleware reports no budget.
retain_history (bool) – To record all the duplicated models even if there are not submitted to the underlying engine. While turning this off might lose part of the submitted model history, it will also reduce the memory cost.
- class nni.nas.strategy.middleware.FailureHandler(*, metric=None, retry_patience=None, failure_types=(ModelStatus.Failed,), retain_history=True)[source]¶
This middleware handles failed models.
The handler supports two modes:
Retry mode: to re-submit the model to the engine, until the model succeeds or patience runs out.
Metric mode: to send a metric for the model, so that the strategy gets penalized for generating this model.
“Failure” doesn’t necessarily mean it has to be the “Failed” state. It can be other types such as “Invalid”, or “Interrupted”, etc. The middleware can thus be chained with other middlewares (e.g.,
Filter), to retry (or put metrics) on invalid models:strategy = Chain( RegularizedEvolution(), FailureHandler(metric=-1.0, failure_types=(ModelStatus.Invalid, )), Filter(filter_fn=custom_constraint) )
- Parameters:
metric (TrialMetric | None) – The metric to send when the model failed. Implies metric mode.
retry_patience (int | None) – Maximum number times of retires. Implies retry mode.
metricandretry_patiencecan’t be both set and can’t be both unset. Exactly one of them must be set.failure_types (tuple[ModelStatus, ...]) – A tuple of
ModelStatus, indicating a set of status that are considered failure.retain_history (bool) – Only has effect in retry mode. If set to
True, submitted models will be kept in a dedicated place, separated from retried models. Otherwise,list_models()might return both submitted models and retried models.
- class nni.nas.strategy.middleware.Filter(filter_fn, metric_for_invalid=None, patience=1000, retain_history=True)[source]¶
Block certain models from submitting.
When models are submitted, they will go through the filter function, to check their validity. If the function returns true, the model will be submitted as usual. Otherwise, the model will be immediately marked as invalid (and optionally have a metric to penalize the strategy).
We recommend to use this middleware to check certain constraints, or prevent the training of some bad models from happening.
- Parameters:
filter_fn (Callable[[ExecutableModelSpace], bool]) – The filter function. Input argument is a
ExecutableModelSpace. ReturningTruemeans the model is good to submit.metric_for_invalid (TrialMetric | None) – When setting to be not None, the metric will be assigned to invalid models. Otherwise, no metric will be set.
patience (int) – Number of continuous invalid models received until the middleware reports no budget.
retain_history (bool) – To faithfully record all the submitted models including the invalid ones. Setting this to false would lose the record of the invalid models, but will also be more memory-efficient. Note that the history can NOT be recovered upon
load_state_dict().
Examples
With
Filter, it becomes a lot easier to have some customized controls for the built-in strategies.For example, if I have a fancy estimator that can tell whether a model’s accuracy is above 90%, and I don’t want any model below 90% submitted for training, I can do:
def some_fancy_estimator(model) -> bool: # return True or False ... strategy = Chain( RegularizedEvolution(), Filter(some_fancy_estimator) )
If the estimator returns false, the model will be immediately marked as invalid, and will not run.
- class nni.nas.strategy.middleware.MedianStop[source]¶
Kill a running model when its best intermediate result so far is worse than the median of results of all completed models at the same number of intermediate reports.
Follow the mechanism in
MedianstopAssessorto stop trials.Warning
This only works theoretically. It can’t be used because engine doesn’t have the ability to kill a model currently.
- class nni.nas.strategy.middleware.MultipleEvaluation(repeat, retain_history=True)[source]¶
Runs each model for multiple times, and use the averaged metric as the final result.
This is useful in scenarios where model evaluation is unstable, with randomness (e.g., Reinforcement Learning).
When models are submitted, replicas of the models will be created (via deepcopy). See
submit_models(). The intermediate metrics, final metric, as well as status will be reduced in their arriving order. For example, the first intermediate metric reported by all replicas will be gathered and averaged, to be the first intermediate metric of the submitted original model. Similar for final metric and status. The status is only considered successful when all the replicas have a successful status. Otherwise, the first unsuccessful status of replicas will be used as the status of the original model.- Parameters:
repeat (int) – How many times to evaluate each model.
retain_history (bool) – If
True, keep all the submitted original models in memory. Otherwiselist_models()will return the replicated models, which, on the other hand saves some memory.
- submit_models(*models)[source]¶
Submit the models.
The method will replicate the models by
repeatnumber of times. If multiple models are submitted simultaneously, the models will be submitted replica by replica. For example, three models are submitted and they are repeated two times, the submitting order will be: model1, model2, model3, model1, model2, model3.Warning
This method might exceed the budget of the underlying engine, even if the budget shows available when the strategy submits.
This method will ignore a model if the model’s replicas is current running.
- class nni.nas.strategy.middleware.StrategyMiddleware(model_space=None, engine=None)[source]¶
StrategyMiddlewareintercepts the models, and strategically filters, mutates, or replicates them and submits them to the engine. It can also intercept the metrics reported by the engine, and manipulates them.StrategyMiddlewareis often used together withChain, which chains a main strategy and a list of middlewares. When a model is created by the main strategy, it is passed to the middlewares in order, during which each middleware have access to the model, and pass it to the next middleware. The metric does quite the opposite, i.e., it is passed from the engine, through all the middlewares, and all the way back to the main strategy.We refer to the middleware closest to the main strategy as upper-level middleware, as it exists at the upper level of the calling stack. Conversely, we refer to the middleware closest to the engine as lower-level middleware.
- property model_space: ExecutableModelSpace¶
Model space is useful for the middleware to do advanced things, e.g., sample its own models.
The model space is set by whoever uses the middleware, before the strategy starts to run.
Utilities¶
- class nni.nas.strategy.utils.DeduplicationHelper(raise_on_dup=False)[source]¶
Helper class to deduplicate samples.
Different from the deduplication on the HPO side, this class simply checks if a sample has been tried before, and does nothing else.
- dedup(sample)[source]¶
If the new sample has not been seen before, it will be added to the history and return True. Otherwise, return False directly.
If raise_on_dup is true, a
DuplicationErrorwill be raised instead of returning False.
- exception nni.nas.strategy.utils.DuplicationError(sample)[source]¶
Exception raised when a sample is duplicated.
- class nni.nas.strategy.utils.RetrySamplingHelper(retries=500, exception_types=(<class 'nni.mutable.exception.SampleValidationError'>, ), raise_last=False)[source]¶
Helper class to retry a function until it succeeds.
Typical use case is to retry random sampling until a non-duplicate / valid sample is found.
- Parameters:
retries (int) – Number of retries.
exception_types (tuple[Type[Exception]]) – Exception types to catch.
raise_last (bool) – Whether to raise the last exception if all retries failed.
One-shot strategies¶
- class nni.nas.strategy.RandomOneShot(filter=None, **kwargs)[source]¶
Train a super-net with uniform path sampling. See reference.
In each step, model parameters are trained after a uniformly random sampling of each choice. Notably, the exporting result is also a random sample of the search space.
The supported mutation primitives of RandomOneShot are:
nni.nas.nn.pytorch.ParametrizedModule(only when parameters’ type is in MutableLinear, MutableConv2d, MutableBatchNorm2d, MutableLayerNorm, MutableMultiheadAttention).
This strategy assumes inner evaluator has set automatic optimization to true.
- Parameters:
filter (ProfilerFilter | dict | Callable[[Sample], bool] | None) – A function that takes a sample and returns a boolean. We recommend using
ProfilerFilterto filter samples. If it’s a dict of keys ofprofiler, and either (or both) ofminandmax, it will be used to construct aRangeProfilerFilter.**kwargs – Parameters for
BaseOneShotStrategy.
Examples
This strategy is mostly used as a “pre”-strategy to speedup another multi-trial strategy. The multi-trial strategy can leverage the trained weights from
RandomOneShotsuch that each sampled model won’t need to be trained from scratch. See SPOS, OFA and AutoFormer for how this is done in the arts.A typical workflow looks like follows:
model_space = MyModelSpace() evaluator = Classification(max_epochs=100) # usually trained longer strategy = RandomOneShot() NasExperiment(model_space, evaluator, strategy).run() # pretrain the supernet # Now the model space is mutated and trained inplace evaluator = Classification(max_epochs=0) # no training strategy = RegularizedEvolution() NasExperiment(model_space, evaluator, strategy).run() # search a subnet
Warning
The second experiment must use
keepmodel format andsequentialexecution engine (which is by default inferred in this setup). Otherwise, the weights will be lost during serialization.For debugging purposes, it’s also possible to save and restore the pretrained supernet:
# After run RandomOneShot strategy torch.save(model_space.state_dict(), '/path/to/somewhere') # Then load the pretrained supernet in a separate run model_space = MyModelSpace() pre_strategy = RandomOneShot() pre_strategy.mutate_model(model_space) model_space.load_state_dict(torch.load('/path/to/somewhere'))
You can also manually use all the methods from
ModelSpacefor the supernet. Notably, thefreeze()method will be weight-preserving, i.e., the weights of the subnet will inherit those on the supernet:model_space.freeze({'layer1': 0, 'layer2': 1})
- class nni.nas.strategy.ENAS(*, batches_per_update=20, log_prob_every_n_step=10, replay_buffer_size=None, reward_metric_name=None, policy_fn=None, update_kwargs=None, warmup_epochs=0, penalty=None, **kwargs)[source]¶
RL controller learns to generate the best network on a super-net. See ENAS paper.
In every epoch, training dataset and validation dataset are given sequentially in batches. For the training dataset, the agent sample subnet from the super-net and train the subnet. For the validation dataset, the agent sample subnet from the super-net and evaluate the subnet; the agent uses the metric evaluated as rewards, put into replay buffer and updates itself.
As the process is similar to the multi-trial version
PolicyBasedRL, this strategy shares some implementations and parameters with it.Attention
ENAS requires the evaluator to report metrics via
self.login itsvalidation_step. See explanation ofreward_metric_namefor details.The supported mutation primitives of ENAS are:
nni.nas.nn.pytorch.InputChoice(only whenn_chosen == 1orn_chosen is None).nni.nas.nn.pytorch.ParametrizedModule(only when parameters are choices and type is in MutableLinear, MutableConv2d, MutableBatchNorm2d, MutableLayerNorm, MutableMultiheadAttention).
Warning
The strategy, under the hood, creates a Lightning module that wraps the Lightning module defined in evaluator, and enables Manual optimization, although we assume the inner evaluator has enabled automatic optimization. We call the optimizers and schedulers configured in evaluator, following the definition in Lightning at best effort, but we make no guarantee that the behaviors are exactly same as automatic optimization. We call
advance_optimization()andadvance_lr_schedulers()to invoke the optimizers and schedulers configured in evaluators. Moreover, some advanced features like gradient clipping will not be supported. If you encounter any issues, please contact us by creating an issue.- Parameters:
batches_per_update (float) – Number of steps for which the gradients will be accumulated, before updating the weights of RL controller.
log_prob_every_n_step (int) – Log the probability of choices every N steps. Useful for visualization and debugging.
replay_buffer_size (int | None) – Size of replay buffer. If it’s none, the size will be the expected trajectory length times
batches_per_update.reward_metric_name (str | None) – The name of the metric which is treated as reward. This will be not effective when there’s only one metric returned from evaluator. If there are multiple, by default, it will find the metric with key name
default. If reward_metric_name is specified, it will find reward_metric_name. Otherwise it raises an exception indicating multiple metrics are found.policy_fn (PolicyFactory | None) – See
PolicyBasedRL.update_kwargs (dict | None) – See
PolicyBasedRL.warmup_epochs (int) – The first
warmup_epochsdo not update architecture weights.penalty (dict | ExpectationProfilerPenalty | SampleProfilerPenalty | None) – If a dict, it should contain the keys:
profiler,baseline, and optionallyscale,nonlinear,aggregate. We will create aSampleProfilerPenaltywith the given parameters. Note that the penalty is operated on the reward, not the loss. Thus in most cases, thescaleshould be set to a negative value.
- class nni.nas.strategy.DARTS(*, arc_learning_rate=0.0003, gradient_clip_val=None, log_prob_every_n_step=10, warmup_epochs=0, penalty=None, **kwargs)[source]¶
Continuous relaxation of the architecture representation, allowing efficient search of the architecture using gradient descent. Reference.
DARTS algorithm is one of the most fundamental one-shot algorithm. DARTS repeats iterations, where each iteration consists of 2 training phases. The phase 1 is architecture step, in which model parameters are frozen and the architecture parameters are trained. The phase 2 is model step, in which architecture parameters are frozen and model parameters are trained. In both phases,
training_stepof the Lightning evaluator will be used.The current implementation corresponds to DARTS (1st order) in paper. Second order (unrolled 2nd-order derivatives) is not supported yet.
Note
DARTS is running a weighted sum of possible architectures under the hood. Please bear in mind that it will be slower and consume more memory that training a single architecture. The common practice is to down-scale the network (e.g., smaller depth / width) for speedup.
New in version 2.8: Supports searching for ValueChoices on operations, with the technique described in FBNetV2: Differentiable Neural Architecture Search for Spatial and Channel Dimensions. One difference is that, in DARTS, we are using Softmax instead of GumbelSoftmax.
The supported mutation primitives of DARTS are:
nni.nas.nn.pytorch.ParametrizedModule(only when parameters are choices and type is in MutableLinear, MutableConv2d, MutableBatchNorm2d, MutableLayerNorm, MutableMultiheadAttention).
Warning
The strategy, under the hood, creates a Lightning module that wraps the Lightning module defined in evaluator, and enables Manual optimization, although we assume the inner evaluator has enabled automatic optimization. We call the optimizers and schedulers configured in evaluator, following the definition in Lightning at best effort, but we make no guarantee that the behaviors are exactly same as automatic optimization. We call
advance_optimization()andadvance_lr_schedulers()to invoke the optimizers and schedulers configured in evaluators. Moreover, some advanced features like gradient clipping will not be supported. If you encounter any issues, please contact us by creating an issue.- Parameters:
arc_learning_rate (float) – Learning rate for architecture optimizer.
gradient_clip_val (float | None) – Clip gradients before optimizing models at each step. Disable gradient clipping by setting it to
None.log_prob_every_n_step (int) – Log current architecture parameters every
log_prob_every_n_stepsteps.warmup_epochs (int) – The first
warmup_epochsdo not update architecture weights.penalty (dict | ExpectationProfilerPenalty | None) – If a dict, it should contain the keys:
profiler,baseline, and optionallyscale,nonlinear,aggregate. We will create aExpectationProfilerPenaltywith the given parameters.**kwargs – Other parameters for
BaseOneShotStrategy.
- class nni.nas.strategy.GumbelDARTS(*, temperature=(1.0, 0.33), **kwargs)[source]¶
Choose the best block by using Gumbel Softmax random sampling and differentiable training. See FBNet and SNAS.
This is a
DARTS-based method that uses gumbel-softmax to simulate one-hot distribution. Essentially, it tries to mimick the behavior of sampling one path on forward by gradually cool down the temperature, aiming to bridge the gap between differentiable architecture weights and discretization of architectures.New in version 2.8: Supports searching for ValueChoices on operations, with the technique described in FBNetV2: Differentiable Neural Architecture Search for Spatial and Channel Dimensions.
The supported mutation primitives of GumbelDARTS are:
nni.nas.nn.pytorch.ParametrizedModule(only when parameters are choices and type is in MutableLinear, MutableConv2d, MutableBatchNorm2d, MutableLayerNorm, MutableMultiheadAttention).
Note
GumbelDARTS is running a weighted sum of possible architectures under the hood. Please bear in mind that it will be slower and consume more memory that training a single architecture. The common practice is to down-scale the network (e.g., smaller depth / width) for speedup.
Warning
The strategy, under the hood, creates a Lightning module that wraps the Lightning module defined in evaluator, and enables Manual optimization, although we assume the inner evaluator has enabled automatic optimization. We call the optimizers and schedulers configured in evaluator, following the definition in Lightning at best effort, but we make no guarantee that the behaviors are exactly same as automatic optimization. We call
advance_optimization()andadvance_lr_schedulers()to invoke the optimizers and schedulers configured in evaluators. Moreover, some advanced features like gradient clipping will not be supported. If you encounter any issues, please contact us by creating an issue.- Parameters:
temperature (dict | tuple[float, float] | LinearTemperatureScheduler | float) –
The temperature used in gumbel-softmax. It can be:
A float, which will be used as the fixed temperature throughout the training.
A tuple of two floats, which will be used as the initial and final temperature for annealing.
A dict with keys
initandmin, which will be used as the initial and final temperature for annealing.A
LinearTemperatureSchedulerinstance.
**kwargs – Other supported parameters can be found in
DARTS.
- class nni.nas.strategy.Proxyless(*, arc_learning_rate=0.0003, gradient_clip_val=None, log_prob_every_n_step=10, warmup_epochs=0, penalty=None, **kwargs)[source]¶
A low-memory-consuming optimized version of differentiable architecture search. See reference.
This is a
DARTS-based method that resamples the architecture to reduce memory consumption. Essentially, it samples one path on forward, and implements its own backward to update the architecture parameters based on only one path.The supported mutation primitives of
Proxylessare:nni.nas.nn.pytorch.LayerChoice(candidate layers must NOT have keyword arguments).nni.nas.nn.pytorch.Repeat(with categorical choice of no transformation).
Warning
The strategy, under the hood, creates a Lightning module that wraps the Lightning module defined in evaluator, and enables Manual optimization, although we assume the inner evaluator has enabled automatic optimization. We call the optimizers and schedulers configured in evaluator, following the definition in Lightning at best effort, but we make no guarantee that the behaviors are exactly same as automatic optimization. We call
advance_optimization()andadvance_lr_schedulers()to invoke the optimizers and schedulers configured in evaluators. Moreover, some advanced features like gradient clipping will not be supported. If you encounter any issues, please contact us by creating an issue.- Parameters:
**kwargs – Supported parameters are the same as
DARTS.
Advanced APIs¶
- class nni.nas.oneshot.pytorch.strategy.OneShotStrategy(mutation_hooks=None, **kwargs)[source]¶
Wrap an one-shot lightning module as a one-shot strategy.
A one-shot strategy has the following workflow:
Mutate the model to a supernet. (The current implementation will do this inplace.)
Mutate the evaluator (must be written in Lightning). Core steps include: injecting the search logics into lightning module and process the dataloaders.
Submit the model and evaluator for training.
Notes
In NNI, we try to separate the “search” part and “training” part in one-shot NAS. The “training” part is defined with evaluator interface (has to be lightning evaluator interface to work with oneshot). Since the lightning evaluator has already broken down the training into minimal building blocks, we can re-assemble them after combining them with the “search” part of a particular algorithm.
After the re-assembling, this module has defined all the search + training. The experiment can use a lightning trainer (which is another part in the evaluator) to train this module, so as to complete the search process.
- Parameters:
mutation_hooks (list[MutationHook] | None) –
Extra mutation hooks to support customized mutation on primitives other than built-ins.
Mutation hooks are callable that inputs an Module and returns a
BaseSuperNetModule. They are invoked intraverse_and_mutate_submodules(), on each submodules. For each submodule, the hook list are invoked subsequently, the later hooks can see the result from previous hooks. The modules that are processed bymutation_hookswill be replaced by the returned module, stored innas_modules, and be the focus of the NAS algorithm.The hook list will be appended by
default_mutation_hooksin each one-shot module.To be more specific, the input arguments of a hook are four arguments:
a module that might be processed,
name of the module in its parent module,
a memo dict whose usage depends on the particular algorithm.
keyword arguments (configurations).
Note that the memo should be read/written by hooks. There won’t be any hooks called on root module.
The returned arguments can be also one of the three kinds:
tuple of:
BaseSuperNetModuleor None, and boolean,boolean,
BaseSuperNetModuleor None.
The boolean value is
suppressindicates whether the following hooks should be called. When it’s true, it suppresses the subsequent hooks, and they will never be invoked. Without boolean value specified, it’s assumed to be false. If a none value appears on the place ofBaseSuperNetModule, it means the hook suggests to keep the module unchanged, and nothing will happen.An example of mutation hook is given in
no_default_hook(). However it’s recommended to implement mutation hooks by derivingBaseSuperNetModule, and add its classmethodmutateto this list.**kwargs (Any) – Extra keyword arguments passed to
Strategy.
- configure_oneshot_module(training_module)[source]¶
Create the oneshot module, i.e., the “search” part of the algorithm.
Subclass should override this.
- list_models(sort=True, limit=1)[source]¶
Getting the best models searched by the one-shot strategy.
The behavior of which models will be chosen depends on the implementation of inner one-shot module.
- Parameters:
sort (bool) – Must be true.
limit (int | None) – The number of models to be returned. Only supports 1 for now.
- mutate_evaluator(evaluator)[source]¶
Mutate the evaluator to the one used in one-shot.
Specifically, it:
uses
oneshot_moduleto wrap themodulein evaluator.calls
preprocess_dataloader()to refuse the dataloaders.
- Return type:
The mutated evaluator.
- mutate_model(model)[source]¶
Convert the model space to a supernet inplace.
The core of a one-shot strategy is usually a carefully-designed supernet, which encodes the sharing pattern and mechanism.
create_supernet()transforms a model space into a one-shot supernet.Mostly useful for debugging and supernet inspection.
- Parameters:
model (ModelSpaceType) – The model space to be transformed. The raw model space written in PyTorch.
- Returns:
The one-shot supernet.
Note that the changes will take inplace.
Therefore the returned model is the same as the input
model.The mutated model is still a
ModelSpaceinstance.In most cases,
simplify()andfreeze(sample)would still return the same result,which is convenient for follow-up search on the supernet.
- Return type:
ModelSpaceType
- property oneshot_module: BaseOneShotLightningModule¶
The one-shot module created by one-shot strategy.
Only available after
run()is called.
- run_hook(hook, name, module, memo)[source]¶
Run a single mutation hook.
For internal use only: subclass can override this to intercept the hooks for customization. For example, provide extra keyword arguments or tamper the memo.
- state_dict()[source]¶
Get the state dict of one-shot strategy.
The state dict of one-shot strategy leverages the checkpoint callback in Lightning evaluator. It will look for
last_model_pathattribute (orbest_model_path) intrainer.checkpoint_callback, save it, and put it back intofit_kwargswhenload_state_dict()is called.
- property supernet: ModelSpace¶
The supernet created by one-shot strategy.
Only available after
run()is called.
- train_dataloader(train_dataloader_fn, val_dataloader_fn)[source]¶
One-shot strategy typically requires fusing train and validation dataloader in an ad-hoc way. As one-shot strategy doesn’t try to open the blackbox of a batch, theoretically, these dataloader can be any dataloader types supported by Lightning.
- Parameters:
train_dataloader_fn (Callable[[], Any]) – Factory that takes no argument, returning a train dataloader.
val_dataloader_fn (Callable[[], Any]) – Similar to
train_dataloader_fn.
- Return type:
Preprocessed train dataloaders.
- val_dataloader(train_dataloader_fn, val_dataloader_fn)[source]¶
See
train_dataloader().- Return type:
Preprocessed validation dataloaders.
base_lightning¶
- class nni.nas.oneshot.pytorch.base_lightning.BaseOneShotLightningModule(training_module)[source]¶
The base class for all one-shot NAS modules.
BaseOneShotLightningModuleis implemented as a subclass ofLightning, to be make it deceptively look like a lightning module to the trainer. It’s actually a wrapper of the lightning module in evaluator. The composition of different lightning modules is as follows:BaseOneShotLightningModule <- Current class (one-shot logics) |_ evaluator.LightningModule <- Part of evaluator (basic training logics) |_ user's model <- Model space, transformed to a supernet by current class.The base class implemented several essential utilities, such as preprocessing user’s model, redirecting lightning hooks for user’s model, configuring optimizers and exporting NAS result are implemented in this class.
- training_module¶
PyTorch lightning module, which defines the training recipe (the lightning module part in evaluator).
- Parameters:
inner_module (pytorch_lightning.LightningModule) – It’s a LightningModule that defines computations, train/val loops, optimizers in a single class. When used in NNI, the
inner_moduleis the combination of instances of evaluator + base model (to be precise, a base model wrapped with LightningModule in evaluator).
- advance_lr_schedulers(batch_idx)[source]¶
Advance the learning rates, when manual optimization is turned on.
The full implementation is here. We only include a partial implementation here. Advanced features like Reduce-lr-on-plateau are not supported.
- advance_optimization(loss, batch_idx, gradient_clip_val=None, gradient_clip_algorithm=None)[source]¶
Run the optimizer defined in evaluators, when manual optimization is turned on.
Call this method when the model should be optimized. To keep it as neat as possible, we only implement the basic
zero_grad,backward,grad_clip, andstephere. Many hooks and pre/post-processing are omitted. Inherit this method if you need more advanced behavior.The full optimizer step could be found here. We only implement part of the optimizer loop here.
- Parameters:
batch_idx (int) – The current batch index.
- architecture_optimizers()[source]¶
Get the optimizers configured in
configure_architecture_optimizers().Return type would be LightningOptimizer or list of LightningOptimizer.
- configure_architecture_optimizers()[source]¶
Hook kept for subclasses. A specific NAS method inheriting this base class should return its architecture optimizers here if architecture parameters are needed. Note that lr schedulers are not supported now for architecture_optimizers.
- Return type:
Optimizers used by a specific NAS algorithm. Return None if no architecture optimizers are needed.
- configure_optimizers()[source]¶
Transparently configure optimizers for the inner model, unless one-shot algorithm has its own optimizer (via
configure_architecture_optimizers()), in which case, the optimizer will be appended to the list.The return value is still one of the 6 types defined in PyTorch-Lightning.
- export()[source]¶
Export the NAS result, ideally the best choice of each
supernet_modules(). You may implement anexportmethod for your customizedsupernet_modules().- Returns:
Keys are labels of mutables, and values are the choice indices of them.
- Return type:
dict
- export_probs()[source]¶
Export the probability of every choice in the search space got chosen.
Note
If such method of some modules is not implemented, they will be simply ignored.
- Returns:
In most cases, keys are labels of the mutables, while values are a dict, whose key is the choice and value is the probability of it being chosen.
- Return type:
dict
- log_probs(probs)[source]¶
Write the probability of every choice to the logger. (nothing related to log-probability stuff).
- Parameters:
probs (Dict[str, Any]) – The result of
export_probs().
- property model: ModelSpace¶
Return the model space defined by the user.
The model space is not guaranteed to have been transformed into a one-shot supernet. For instance, when
__init__hasn’t completed, the model space will still be the original one.
- postprocess_weight_optimizers(optimizers)[source]¶
Some subclasss need to modify the original optimizers. This is where it should be done. For example, differentiable algorithms might not want the architecture weights to be inside the weight optimizers.
- Return type:
By default, it return the original object.
- resample()[source]¶
Trigger the resample for each
supernet_modules(). Sometimes (e.g., in differentiable cases), it does nothing.- Returns:
Sampled architecture.
- Return type:
dict
- class nni.nas.oneshot.pytorch.base_lightning.BaseSuperNetModule(*args, **kwargs)[source]¶
Mutated module in super-net. Usually, the feed-forward of the module itself is undefined. It has to be resampled with
resample()so that a specific path is selected. (Sometimes, this is not required. For example, differentiable super-net.)A super-net module usually corresponds to one sample. But two exceptions:
A module can have multiple parameter spec. For example, a convolution-2d can sample kernel size, channels at the same time.
Multiple modules can share one parameter spec. For example, multiple layer choices with the same label.
For value choice compositions, the parameter spec are bounded to the underlying (original) value choices, rather than their compositions.
- export(memo)[source]¶
Export the final architecture within this module. It should have the same keys as
search_space_spec().- Parameters:
memo (dict[str, Any]) – Use memo to avoid the same label gets exported multiple times.
- export_probs(memo)[source]¶
Export the probability / logits of every choice got chosen.
- Parameters:
memo (dict[str, Any]) – Use memo to avoid the same label gets exported multiple times.
- classmethod mutate(module, name, memo, mutate_kwargs)[source]¶
This is a mutation hook that creates a
BaseSuperNetModule. The method should be implemented in each specific super-net module, because they usually have specific rules about what kind of modules to operate on.- Parameters:
module (nn.Module) – The module to be mutated (replaced).
name (str) – Name of this module. With full prefix. For example,
module1.block1.conv.memo (dict) – Memo to enable sharing parameters among mutated modules. It should be read and written by mutate functions themselves.
mutate_kwargs (dict) – Algo-related hyper-parameters, and some auxiliary information.
- Returns:
The mutation result, along with an optional boolean flag indicating whether to suppress follow-up mutation hooks. See
BaseOneShotLightningModulefor details.- Return type:
Union[BaseSuperNetModule, bool, tuple[BaseSuperNetModule, bool]]
supermodule.differentiable¶
- class nni.nas.oneshot.pytorch.supermodule.differentiable.DifferentiableMixedCell(*args, **kwargs)[source]¶
Implementation of Cell under differentiable context.
Similar to PathSamplingCell, this cell only handles cells of specific kinds (e.g., with loose end).
An architecture parameter is created on each edge of the full-connected graph.
- class nni.nas.oneshot.pytorch.supermodule.differentiable.DifferentiableMixedInput(*args, **kwargs)[source]¶
Mixed input. Forward returns a weighted sum of candidates. Implementation is very similar to
DifferentiableMixedLayer.- Parameters:
n_candidates (int) – Expect number of input candidates.
n_chosen (int) – Expect numebr of inputs finally chosen.
alpha (Tensor) – Tensor that stores the “learnable” weights.
softmax (nn.Module) – Customizable softmax function. Usually
nn.Softmax(-1).label (str) – Name of the choice.
- class nni.nas.oneshot.pytorch.supermodule.differentiable.DifferentiableMixedLayer(*args, **kwargs)[source]¶
Mixed layer, in which fprop is decided by a weighted sum of several layers. Proposed in DARTS: Differentiable Architecture Search.
The weight
alphais usually learnable, and optimized on validation dataset.Differentiable sampling layer requires all operators returning the same shape for one input, as all outputs will be weighted summed to get the final output.
- Parameters:
paths (list[tuple[str, nn.Module]]) – Layers to choose from. Each is a tuple of name, and its module.
alpha (Tensor) – Tensor that stores the “learnable” weights.
softmax (nn.Module) – Customizable softmax function. Usually
nn.Softmax(-1).label (str) – Name of the choice.
- class nni.nas.oneshot.pytorch.supermodule.differentiable.DifferentiableMixedRepeat(*args, **kwargs)[source]¶
Implementation of Repeat in a differentiable supernet. Result is a weighted sum of possible prefixes, sliced by possible depths.
If the output is not a single tensor, it will be summed at every independant dimension. See
weighted_sum()for details.
- class nni.nas.oneshot.pytorch.supermodule.differentiable.GumbelSoftmax(dim=-1)[source]¶
Wrapper of
F.gumbel_softmax. dim = -1 by default.
- class nni.nas.oneshot.pytorch.supermodule.differentiable.MixedOpDifferentiablePolicy(operation, memo, mutate_kwargs)[source]¶
Implements the differentiable sampling in mixed operation.
One mixed operation can have multiple value choices in its arguments. Thus the
_arch_alphahere is a parameter dict, andnamed_parametersfilters out multiple parameters with_arch_alphaas its prefix.When this class is asked for
forward_argument, it returns a distribution, i.e., a dict from int to float based on its weights.All the parameters (
_arch_alpha,parameters(),_softmax) are saved as attributes ofoperation, rather thanself, because this class itself is not ann.Module, and saved parameters here won’t be optimized.
supermodule.sampling¶
- class nni.nas.oneshot.pytorch.supermodule.sampling.MixedOpPathSamplingPolicy(operation, memo, mutate_kwargs)[source]¶
Implements the path sampling in mixed operation.
One mixed operation can have multiple value choices in its arguments. Each value choice can be further decomposed into “leaf value choices”. We sample the leaf nodes, and composits them into the values on arguments.
- class nni.nas.oneshot.pytorch.supermodule.sampling.PathSamplingCell(*args, **kwargs)[source]¶
The implementation of super-net cell follows DARTS.
When
factory_usedis true, it reconstructs the cell for every possible combination of operation and input index, because for different input index, the cell factory could instantiate different operations (e.g., with different stride). On export, we first have best (operation, input) pairs, the select the bestnum_ops_per_node.loose_endis not supported yet, because it will cause more problems (e.g., shape mismatch). We assumesloose_endto beallregardless of its configuration.A supernet cell can’t slim its own weight to fit into a sub network, which is also a known issue.
- class nni.nas.oneshot.pytorch.supermodule.sampling.PathSamplingInput(*args, **kwargs)[source]¶
Mixed input. Take a list of tensor as input, select some of them and return the sum.
- _sampled¶
Sampled input indices.
- Type:
int or list of int
- class nni.nas.oneshot.pytorch.supermodule.sampling.PathSamplingLayer(*args, **kwargs)[source]¶
Mixed layer, in which fprop is decided by exactly one inner layer or sum of multiple (sampled) layers. If multiple modules are selected, the result will be summed and returned.
- _sampled¶
Sampled module indices.
- Type:
int or list of str
- label¶
Name of the choice.
- Type:
str
supermodule.proxyless¶
Implementation of ProxylessNAS: a hyrbid approach between differentiable and sampling. The support remains limited. Known limitations include:
No support for multiple arguments in forward.
No support for mixed-operation (value choice).
The code contains duplicates. Needs refactor.
- class nni.nas.oneshot.pytorch.supermodule.proxyless.ProxylessMixedInput(*args, **kwargs)[source]¶
Proxyless version of differentiable input choice. See
ProxylessMixedLayerfor implementation details.- export(memo)[source]¶
Same as
resample().
- class nni.nas.oneshot.pytorch.supermodule.proxyless.ProxylessMixedLayer(*args, **kwargs)[source]¶
Proxyless version of differentiable mixed layer. It resamples a single-path every time, rather than compute the weighted sum.
Currently the input and output of the candidate layers can only be tensors or tuple of tensors. They can’t be dict, list or any complex types, or non-tensors (including none).
- export(memo)[source]¶
Same as
resample().
- class nni.nas.oneshot.pytorch.supermodule.proxyless.ProxylessMixedRepeat(*args, **kwargs)[source]¶
ProxylessNAS converts repeat to a sequential blocks of layer choices between the original block and an identity layer.
Only pure categorical depth choice is supported. If the categorical choices are not consecutive integers, the constraint will only be considered at export.
supermodule.operation¶
Operations that support weight sharing at a fine-grained level, which is commonly known as super-kernel (as in channel search), or weight entanglement.
- class nni.nas.oneshot.pytorch.supermodule.operation.MixedBatchNorm2d(*args, **kwargs)[source]¶
Mixed BatchNorm2d operation.
Supported arguments are:
num_featureseps(only supported in path sampling)momentum(only supported in path sampling)
For path-sampling, prefix of
weight,bias,running_meanandrunning_varare sliced. For weighted cases, the maximumnum_featuresis used directly.Momentum is required to be float. PyTorch BatchNorm supports a case where momentum can be none, which is not supported here.
- class nni.nas.oneshot.pytorch.supermodule.operation.MixedConv2d(*args, **kwargs)[source]¶
Mixed conv2d op.
Supported arguments are:
in_channelsout_channelsgroupsstride(only supported in path sampling)kernel_sizepaddingdilation(only supported in path sampling)
paddingwill be the “max” padding in differentiable mode.Mutable
groupsis NOT supported in most cases of differentiable mode. However, we do support one special case when the group number is proportional toin_channelsandout_channels. This is often the case of depth-wise convolutions.For channels, prefix will be sliced. For kernels, we take the small kernel from the center and round it to floor (left top). For example
max_kernel = 5*5, sampled_kernel = 3*3, then we take [1: 4] max_kernel = 5*5, sampled_kernel = 2*2, then we take [1: 3] □ □ □ □ □ □ □ □ □ □ □ ■ ■ ■ □ □ ■ ■ □ □ □ ■ ■ ■ □ □ ■ ■ □ □ □ ■ ■ ■ □ □ □ □ □ □ □ □ □ □ □ □ □ □ □ □
- class nni.nas.oneshot.pytorch.supermodule.operation.MixedLayerNorm(*args, **kwargs)[source]¶
Mixed LayerNorm operation.
Supported arguments are:
normalized_shapeeps(only supported in path sampling)
For path-sampling, prefix of
weightandbiasare sliced. For weighted cases, the maximumnormalized_shapeis used directly.eps is required to be float.
- class nni.nas.oneshot.pytorch.supermodule.operation.MixedLinear(*args, **kwargs)[source]¶
Mixed linear operation.
Supported arguments are:
in_featuresout_features
Prefix of weight and bias will be sliced.
- class nni.nas.oneshot.pytorch.supermodule.operation.MixedMultiHeadAttention(*args, **kwargs)[source]¶
Mixed multi-head attention.
Supported arguments are:
embed_dimnum_heads(only supported in path sampling)kdimvdimdropout(only supported in path sampling)
At init, it constructs the largest possible Q, K, V dimension. At forward, it slices the prefix to weight matrices according to the sampled value. For
in_proj_biasandin_proj_weight, three parts will be sliced and concatenated together:[0, embed_dim),[max_embed_dim, max_embed_dim + embed_dim),[max_embed_dim * 2, max_embed_dim * 2 + embed_dim).Warning
All candidates of
embed_dimshould be divisible by all candidates ofnum_heads.
- class nni.nas.oneshot.pytorch.supermodule.operation.MixedOperation(*args, **kwargs)[source]¶
This is the base class for all mixed operations. It’s what you should inherit to support a new operation with mutable.
It contains commonly used utilities that will ease the effort to write customized mixed operations, i.e., operations with mutable in its arguments. To customize, please write your own mixed operation, and add the hook into
mutation_hooksparameter when using the strategy.By design, for a mixed operation to work in a specific algorithm, at least two classes are needed.
One class needs to inherit this class, to control operation-related behavior, such as how to initialize the operation such that the sampled operation can be its sub-operation.
The other one needs to inherit
MixedOperationSamplingPolicy, which controls algo-related behavior, such as sampling.
The two classes are linked with
sampling_policyattribute inMixedOperation, whose type is set viamixed_op_samplinginmutate_kwargswhenMixedOperation.mutate()is called.With this design, one mixed-operation (e.g., MixedConv2d) can work in multiple algorithms (e.g., both DARTS and ENAS), saving the engineering effort to rewrite all operations for each specific algo.
This class should also define a
bound_type, to control the matching type in mutate, anargument_list, to control which arguments can be dynamically used inforward. This list will also be used in mutate for sanity check.- export(memo)[source]¶
Delegates to
MixedOperationSamplingPolicy.export().
- export_probs(memo)[source]¶
Delegates to
MixedOperationSamplingPolicy.export_probs().
- forward(*args, **kwargs)[source]¶
First get sampled arguments, then forward with the sampled arguments (by calling
forward_with_args).
- forward_argument(name)[source]¶
Get the argument used in forward. This if often related to algo. We redirect this to sampling policy.
- forward_with_args(*args, **kwargs)[source]¶
To control real fprop. The accepted arguments are
argument_list, appended by forward arguments in thebound_type.
- freeze(sample)[source]¶
Freeze the mixed operation to a specific operation. Weights will be copied from the mixed operation to the frozen operation.
The returned operation will be of the
bound_type.
- freeze_weight(**kwargs)[source]¶
Slice the params and buffers for subnet forward and state dict.
The arguments are same as the arguments passed to
__init__.
- classmethod mutate(module, name, memo, mutate_kwargs)[source]¶
Find value choice in module’s arguments and replace the whole module
- resample(memo)[source]¶
Delegates to
MixedOperationSamplingPolicy.resample().
- super_init_argument(name, value_choice)[source]¶
Get the initialization argument when constructing super-kernel, i.e., calling
super().__init__(). This is often related to specific operator, rather than algo.For example:
def super_init_argument(self, name, value_choice): return max(value_choice.grid())
- class nni.nas.oneshot.pytorch.supermodule.operation.MixedOperationSamplingPolicy(operation, memo, mutate_kwargs)[source]¶
Algo-related part for mixed Operation.
MixedOperationdelegates its resample and export to this policy (or its subclass), so that one Operation can be easily combined with different kinds of sampling.One SamplingStrategy corresponds to one mixed operation.
- export(operation, memo)[source]¶
The handler of
MixedOperation.export().
- export_probs(operation, memo)[source]¶
The handler of
MixedOperation.export_probs().
- forward_argument(operation, name)[source]¶
Computing the argument with
nameused in operation’s forward. Usually a value, or a distribution of value.
- resample(operation, memo)[source]¶
The handler of
MixedOperation.resample().
Profiler Utilities¶
Guide the one-shot strategy to sample architecture within a target latency.
This module converts the profiling results returned by profiler to something that one-shot strategies can understand. For example, a loss or some penalty to the reward.
This file is experimentally placed in the oneshot package. It might be moved to a more general place in the future.
- class nni.nas.oneshot.pytorch.profiler.ExpectationProfilerPenalty(profiler, baseline, scale=1.0, *, nonlinear='linear', aggregate='add')[source]¶
ProfilerPenaltyfor a sample with distributions. Value for each label is a a mapping from chosen value to probablity.
- class nni.nas.oneshot.pytorch.profiler.ProfilerFilter(profiler)[source]¶
Filter the sample based on the result of the profiler.
Subclass should implement the
filtermethod that returns true or false to indicate whether the sample is valid.Directly call the instance of this class will call the
filtermethod.
- class nni.nas.oneshot.pytorch.profiler.ProfilerPenalty(profiler, baseline, scale=1.0, *, nonlinear='linear', aggregate='add')[source]¶
Give the loss a penalty with the result on the profiler.
Latency losses in TuNAS and ProxylessNAS are its special cases.
The computation formula is divided into two steps, where we first compute a
normalized_penalty, whose zero point is when the penalty meets the baseline, and then we aggregate it with the original loss.\[\begin{split}\begin{aligned} \text{normalized_penalty} ={} & \text{nonlinear}(\frac{\text{penalty}}{\text{baseline}} - 1) \\ \text{loss} ={} & \text{aggregate}(\text{original_loss}, \text{normalized_penalty}) \end{aligned}\end{split}\]where
penaltyhere is the result returned by the profiler.For example, when
nonlinearispositiveandaggregateisadd, the computation formula is:\[\text{loss} = \text{original_loss} + \text{scale} * (max(\frac{\text{penalty}}{\text{baseline}}, 1) - 1, 0)\]- Parameters:
profiler (Profiler) – The profiler which is used to profile the sample.
scale (float) – The scale of the penalty.
baseline (float) – The baseline of the penalty.
nonlinear (Literal['linear', 'positive', 'negative', 'absolute']) – The nonlinear function to apply to \(\frac{\text{penalty}}{\text{baseline}}\). The result is called
normalized_penalty. Iflinear, then keep the original value. Ifpositive, then apply the function \(max(0, \cdot)\). Ifnegative, then apply the function \(min(0, \cdot)\). Ifabsolute, then apply the function \(abs(\cdot)\).aggregate (Literal['add', 'mul']) – The aggregate function to merge the original loss with the penalty. If
add, then the final loss is \(\text{original_loss} + \text{scale} * \text{normalized_penalty}\). Ifmul, then the final loss is \(\text{original_loss} * (1 + \text{normalized_penalty})^{\text{scale}}\).
- class nni.nas.oneshot.pytorch.profiler.RangeProfilerFilter(profiler, min=None, max=None)[source]¶
Give up the sample if the result of the profiler is out of range.
minandmaxcan’t be both None.- Parameters:
profiler (Profiler) – The profiler which is used to profile the sample.
min (float | None) – The lower bound of the profiler result. None means no minimum.
max (float | None) – The upper bound of the profiler result. None means no maximum.
- class nni.nas.oneshot.pytorch.profiler.SampleProfilerPenalty(profiler, baseline, scale=1.0, *, nonlinear='linear', aggregate='add')[source]¶
ProfilerPenaltyfor a single sample. Value for each label is a specifically chosen value.
Experiment¶
- class nni.nas.experiment.NasExperiment(model_space, evaluator, strategy, config=None, id=None)[source]¶
The entry for a NAS experiment. Users can use this class to start/stop or inspect an experiment, like exporting the results.
Experiment is a sub-class of
nni.experiment.Experiment, there are many similarities such as configurable training service to distributed running the experiment on remote server. But unlikenni.experiment.Experiment,NasExperimentdoesn’t support configure:trial_code_directory, which can only be current working directory.search_space, which is auto-generated in NAS.trial_command, which is auto-set to launch the modulized trial code.
NasExperimentalso doesn’t have tuner/assessor/advisor, because such functionality is already implemented in strategy.Also, unlike
nni.experiment.Experimentwhich is bounded to a node server,NasExperimentoptionally starts a node server to schedule the trials, depending on the configuration of execution engine. When the strategy is one-shot, the step of launching node server is omitted, and the experiment is run locally by default.Configurations of experiments, such as execution engine, number of GPUs allocated, should be put into a
NasExperimentConfigand passed to the initialization of an experiment. The config can be also altered after the experiment is initialized.- Parameters:
model_space (BaseModelSpace) – The model space to search.
evaluator (Evaluator) – Evaluator for the experiment.
strategy (Strategy) – Exploration strategy. Can be multi-trial or one-shot.
config (nni.nas.experiment.config.experiment.NasExperimentConfig) – Configurations of the experiment. See
NasExperimentConfigfor details. When not provided, a default config will be created based on current model space, evaluator and strategy. Detailed rules can be found innni.nas.experiment.NasExperimentConfig.default().
Warning
wait_completiondoesn’t work for NAS experiment because NAS experiment always wait for completion.Examples
>>> base_model = MobileNetV3Space() >>> search_strategy = strategy.Random() >>> model_evaluator = Classification() >>> exp = NasExperiment(base_model, model_evaluator, search_strategy) >>> exp_config.max_trial_number = 20 >>> exp_config.training_service.use_active_gpu = False >>> exp.run(exp_config, 8081)
Export top models and re-initialize the top model:
>>> for model_dict in exp.export_top_models(formatter='dict'): ... print(model_dict) >>> with model_context(model_dict): ... final_model = Net()
- export_top_models(top_k=1, *, formatter=None, **kwargs)[source]¶
Export several top performing models.
The concrete behavior of export depends on each strategy. See the documentation of each strategy for detailed specifications.
- Parameters:
top_k (int) – How many models are intended to be exported.
formatter (Literal['code', 'dict', 'instance'] | None) –
If formatter is none, original
ExecutableModelSpaceobjects will be returned. Otherwise, the formatter will be used to convert the model space to a human-readable format. The formatter could be:code: the python code of model will be returned (only forGraphModelSpace).dict: the sample (architecture dict) that is used to freeze the model space.instance: the instantiated callable model.
- load_checkpoint()[source]¶
Recover the status of an experiment from checkpoint.
It first loads the config, and then loads status for strategy and engine. The config must match exactly with the config used to create this experiment.
The status of strategy and engine will only be loaded if engine has been created, and the checkpoint file exists.
Notes
This method is called twice when loading an experiment:
When resume is just called, the config will be loaded and will be cross-checked with the current config.
After NNI manager is started and engine is created, the full method is called to load the state of strategy and engine. For this time, the config will be loaded and cross-checked again.
Semantically, “loading config” and “loading status” are two different things which should be done separately. The current implementation is a bit hacky, but it’s simple and works.
- load_state_dict(state_dict)[source]¶
Load the state dict to recover the status of experiment.
NOTE: This should only be called after the engine is created (i.e., after calling
start()).
- save_checkpoint()[source]¶
Save the whole experiment state.
It will dump the config first (as a JSON) and then states of components like strategy and engine. It calls
state_dict()to get the states.
- start(port=8080, debug=False, run_mode=RunMode.Background)[source]¶
Start a NAS experiment.
Since NAS experiments always have strategies running in main thread,
start()will not exit until the strategy finishes its run.portandrun_modeare only meaningful when_nni_manager_required()returns true.- Parameters:
port (int) – Port to start NNI manager.
debug (bool) – If true, logging will be in debug mode.
run_mode (RunMode) – Whether to have the NNI manager in background, or foreground.
See also
- class nni.nas.experiment.config.CgoEngineConfig(name=None, **kwargs)[source]¶
Engine for cross-graph optimization.
- class nni.nas.experiment.config.ExecutionEngineConfig(name=None, **kwargs)[source]¶
Base class for execution engine config. Useful for instance check.
- class nni.nas.experiment.config.GraphModelFormatConfig(name=None, **kwargs)[source]¶
Model format config for graph-based model space.
- class nni.nas.experiment.config.ModelFormatConfig(name=None, **kwargs)[source]¶
Base class for model format config. Useful for instance check.
- class nni.nas.experiment.config.NasExperimentConfig(*args, **kwargs)[source]¶
Config for NAS experiment.
Other than training service fields which also exists in a HPO experiment, additional fields provided by NAS include execution engine and model format. Execution engine is used to specify how to (e.g., distributedly or sequentially) run a trial, and model format specifies the format of the converted model space used throughout the NAS experiment.
It can be constructed via 3 approaches.
Create a default config and then modify some fields (recommended). The default config should be good enough for most cases. Users only need to update some fields like concurrency. See details in
default(). Example:config = NasExperimentConfig.default(model_space, evaluator, strategy) config.trial_concurrency = 4
Create an object by providing several required fields, and then set other fields. Though marked as optional in function signature, it’s recommended to set all three fields.
config = NasExperimentConfig(‘ts’, ‘graph’, ‘local’) config.experiment_name = ‘hello’ config.execution_engine.dummy_input = [1, 3, 224, 224]
Create an empty object and set all fields manually. Put the fields into kwargs should also work:
config = NasExperimentConfig() config.execution_engine = TrainingServiceEngineConfig() config.model_format = SimplifiedModelFormatConfig() config.training_service = LocalConfig(use_active_gpu=True) # equivalent to config = NasExperimentConfig( execution_engine=TrainingServiceEngineConfig(), model_format=SimplifiedModelFormatConfig(), training_service=LocalConfig(use_active_gpu=True) )
- classmethod default(model_space, evaluator, strategy)[source]¶
Instantiate a default config. Infer from current setting of model space, evaluator and strategy.
If the strategy is found to be a one-shot strategy, the execution engine will be set to “sequential” and model format will be set to “raw” to preserve the weights and the model object.
If the strategy is found to be a multi-trial strategy, training service engine will be used by default, and the training service will be set to “local” if not provided. Model format will be set to “simplified” for performance and memory efficiency.
- class nni.nas.experiment.config.RawModelFormatConfig(name=None, **kwargs)[source]¶
Model format that keeps the original model space.
- class nni.nas.experiment.config.SequentialEngineConfig(name=None, **kwargs)[source]¶
Engine that executes the models sequentially.
Profiler¶
- class nni.nas.profiler.Profiler(model_space)[source]¶
Profiler is a class that profiles the performance of a model within a space.
Unlike the regular profilers, NAS profilers are initialized with a space, and are expected to do some pre-computation with the space, such that it can quickly computes the performance of a model given a sample within a space.
A profiler can return many things, such as latency, throughput, model size, etc. Mostly things that can be computed instantly, or can be computed with a small overhead. For metrics that require training, please use
Evaluatorinstead.
- class nni.nas.profiler.ExpressionProfiler(model_space)[source]¶
Profiler whose
profile()method is an evaluation of a precomputed expression.This type of profiler is useful for optimization and analysis. For example, to find the best model size is equivalent to find the minimum value of the expression. Users can also compute the mathematical expression for a distribution of model samples.
FLOPs¶
- class nni.nas.profiler.pytorch.flops.FlopsParamsCounterConfig(count_bias=True, count_normalization=True, count_activation=True)[source]¶
Configuration for counting FLOPs.
- count_bias¶
Whether to count bias into FLOPs.
- Type:
bool
- count_normalization¶
Whether to count normalization (e.g., Batch normalization) into FLOPs and parameters.
- Type:
bool
- count_activation¶
Whether to count activation (e.g., ReLU) into FLOPs.
- Type:
bool
- class nni.nas.profiler.pytorch.flops.FlopsParamsProfiler(model_space, args, **kwargs)[source]¶
The profiler to count flops and parameters of a model.
It first runs shape inference on the model to get the input/output shapes of all the submodules. Then it traverse the submodules and use registered formulas to count the FLOPs and parameters as an expression. The results are stored in a
FlopsResultobject. When a sample is provided, the expressions are frozen and the results are computed.Notes
Customized FLOPs formula can be registered by using
register_flops_formula(). It takes three mandatory arguments: the module itself, input shapes as a tuple ofMutableShapeobjects, and output shapes as a tuple ofMutableShapeobjects. It also takes some additional keyword arguments:name: the name of the module in the PyTorch module hierarchy.shapes: a dictionary of all the input and output shapes of all the modules.config: the configuration object ofFlopsParamsProfiler.
If fields in
FlopsParamsCounterConfigare used in the formula, they will also be passed as keyword arguments.It then returns a
FlopsResultobject that contains the FLOPs and parameters of the module.For example, to count the FLOPs of a unbiased linear layer, we can register the following formula:
def linear_flops(module, input_shape, output_shape, *, name, shapes, config): x, y = input_shape[0], output_shape[0] # unpack the tuple return FlopsResult( flops=x[1:].numel() * module.out_features, # forget the batch size params=module.in_features * module.out_features ) register_flops_formula(nn.Linear, linear_flops)
- Parameters:
model_space (ModelSpace) – The model space to profile.
args (Any) – Dummy inputs to the model to count flops. Similar to torch.onnx.export, the input can be a tensor or a tuple of tensors, or a tuple of arguments ends with a dictionary of keyword arguments.
**kwargs – Additional configurations. See
FlopsParamsCounterConfigfor supported arguments.
- class nni.nas.profiler.pytorch.flops.FlopsProfiler(model_space, args, **kwargs)[source]¶
The FLOPs part of
FlopsParamsProfiler.Batch size is not considered (actually ignored on purpose) in flops profiling.
- Parameters:
model_space (ModelSpace) – The model space to profile.
args (Any) – Dummy inputs to the model to count flops. Similar to torch.onnx.export, the input can be a tensor or a tuple of tensors, or a tuple of arguments ends with a dictionary of keyword arguments.
**kwargs (Any) – Additional configurations. See
FlopsParamsCounterConfigfor supported arguments.
- class nni.nas.profiler.pytorch.flops.NumParamsProfiler(model_space, args, **kwargs)[source]¶
The parameters part of
FlopsParamsProfiler.- Parameters:
model_space (ModelSpace) – The model space to profile.
args (Any) – Dummy inputs to the model to count flops. Similar to torch.onnx.export, the input can be a tensor or a tuple of tensors, or a tuple of arguments ends with a dictionary of keyword arguments.
**kwargs (Any) – Additional configurations. See
FlopsParamsCounterConfigfor supported arguments.
- nni.nas.profiler.pytorch.flops.count_flops_params(name, module, shapes, config)[source]¶
Count FLOPs of a module.
Firstly check whether the type of module is in FLOPs formula registry. If not, traverse its children and sum up the FLOPs of each child.
- Parameters:
name (str) – Name of the module.
module (Module) – The module to count FLOPs.
shapes (dict[str, tuple[MutableShape, MutableShape]]) – Input and output shapes of all the modules. Should at least contain
name.
- Returns:
The FLOPs of the module.
- Return type:
flops
- nni.nas.profiler.pytorch.flops.register_flops_formula(module_type, formula)[source]¶
Register a FLOPs counting formula for a module.
- Parameters:
module_type (Any) – The module type to register the formula for. The class here needs to be a class, not an instantiated module.
formula (Callable[[...], FlopsResult]) – A function that takes in a module and its inputs, and returns
FlopsResult. CheckFlopsParamsProfilerfor more details.
nn-Meter¶
- class nni.nas.profiler.pytorch.nn_meter.NnMeterProfiler(model_space, args, predictor, custom_leaf_types=None, simplify_shapes=False)[source]¶
Profiler based on nnMeter, which is a tool to estimate the latency of neural networks without real device.
The profiler breaks the whole model into submodules and profiles each of them, introducing branches when some part of the model contains mutables. The latency of a module is the sum of the latency of its submodules.
NnMeterProfilerdoes not respectis_leaf_module()when it profiles the latency of the model space. To control the granularity, inherit this class and overrideis_leaf_module().- Parameters:
model_space (ModelSpace) – The model space to profile.
args (Any) – Dummy inputs to the model to count flops. Similar to torch.onnx.export, the input can be a tensor or a tuple of tensors, or a tuple of arguments ends with a dictionary of keyword arguments.
predictor (str | nnMeterPredictor) – The latency predictor to use. Can be a string (alias of nnMeterPredictor) or a
nnMeterPredictor.custom_leaf_types (tuple[type, ...] | None) – A tuple of types of modules that should be considered as leaf modules.
simplify_shapes (bool) – Experimental feature. If True, the shapes of the inputs and outputs of each module will be mathematically simplified with the underlying sympy library.
- estimate_latency(name, module, shapes)[source]¶
Count the latency of a mutable module with the given mutable input shapes.
Returns a mutable expression that is the template of the latency.
- Parameters:
name (str) – The name of the module.
module (nn.Module) – The module to count latency.
shapes (dict[str, Any]) – The input shapes to the module.
- estimate_layerchoice_latency(name, module, shapes)[source]¶
Estimate the latency of a layer choice.
Profile each choice block and merge them into a switch-case expression.
- estimate_repeat_latency(name, module, shapes)[source]¶
Estimate the latency of a Repeat.
Profile each block and merge possibilities at different depths into a switch-case expression.
- is_leaf_module(module)[source]¶
If this method returns true for a module, the profiler will exhaust all the possible freeze result of the module, and gets each latency respectively.
By default, it returns true for modules where
is_leaf_module()returns true, or forMutableModulebut not aLayerChoiceorRepeator a model space without dangling mutables.
- nni.nas.profiler.pytorch.nn_meter.combinations(module, input_shape)[source]¶
List all the combinations of the (mutable) module and the input shape.
The returned iterator yields a tuple of (sample, module, input) for each combination. The inputs will be generated with
torch.randn()based on the sampled input shape.The module can be potentially not any mutable object. If the module is not a
Mutable, it must be ann.Moduleso that it can be wrapped with a MutableModule.
- nni.nas.profiler.pytorch.nn_meter.sample_to_condition(mutables, sample)[source]¶
Convert a sample to a condition that can be used to verify whether a new sample is compatible with the old one. Freeze the returned condition with a certain sample to get a boolean value.
- Parameters:
mutables (dict[str, LabeledMutable]) – A dictionary mapping label to mutable. Get it from
Mutable.simplify().sample (Sample) – A sample to convert.
Model format¶
- class nni.nas.space.BaseModelSpace[source]¶
A model space is a collection of mutables, organized in a meaningful way (i.e., in a model way).
BaseModelSpaceis almost only used for isinstance check. A few utility functions might be provided inside this class for convenience.
- class nni.nas.space.Edge(head, tail, _internal=False)[source]¶
A tensor, or “data flow”, between two nodes.
Example forward code snippet:
a, b, c = split(x) p = concat(a, c) q = sum(b, p) z = relu(q)
Edges in above snippet:
+ head: (split, 0), tail: (concat, 0) # a in concat + head: (split, 2), tail: (concat, 1) # c in concat + head: (split, 1), tail: (sum, -1 or 0) # b in sum + head: (concat, null), tail: (sum, -1 or 1) # p in sum + head: (sum, null), tail: (relu, null) # q in relu
- graph¶
Graph.
- head¶
Head node.
- tail¶
Tail node.
- head_slot¶
Index of outputs in head node. If the node has only one output, this should be
null.
- tail_slot¶
Index of inputs in tail node. If the node has only one input, this should be
null. If the node does not care about order, this can be-1.
- class nni.nas.space.ExecutableModelSpace(status=ModelStatus.Initialized)[source]¶
Model space with an extra execute method that defines how the models should be evaluated. It should be
ModelSpaceWithExecutionbut that’s too long.Both model space, as well as single models mutated from the space, will be instances of
ExecutableModelSpace. They only differ in the status flag (seeModelStatus).Since the single models that are directly evaluated are also of this type, this class has an
execute()method which defines how the training pipeline works, i.e., how to assemble the evaluator and the model, and how to execute the training and evaluation.By convention, only frozen models (status is
ModelStatus.Frozen) and instances ofExecutableModelSpacecan be sent to execution engine for training.In most cases,
ExecutableModelSpaceonly contains the necessary information that is required for NAS mutations and reconstruction of the original model. This makes the model space light-weighted, and easy to be serialized for sending to clusters. It also reforms the space to be more friendly to NAS algorithms (e.g., in the format of graphs).- executable_model()[source]¶
Fully instantiate the deep learning model (e.g., PyTorch Module) so that it’s ready to be executed.
executable_model()is usually symmetrical tofrom_model(). Whilefrom_model()converts deep learning model toExecutableModelSpace,executable_model()convertsExecutableModelSpaceback to deep learning model.- Returns:
Typical this method should return a PyTorch / Tensorflow model (or model factory),
depending on the input format of evaluator.
- Return type:
Any
- classmethod from_model(model_space, evaluator=None, **configs)[source]¶
Convert any model space to a specific type of executable model space.
- Parameters:
model_space (BaseModelSpace) – Model space written in deep learning framework in most cases.
evaluator (Evaluator | None) – A model usually requires an evaluator to be executable. But evaluator can sometimes be optional for debug purposes or to support fancy algorithms.
configs (Any) – Additional configurations for the executable model space.
- Return type:
The converted model space.
- property metric: TrialMetric | None¶
Training result of the model, or
Noneif it’s not yet trained or has failed to train.
- sample: Sample | None¶
The sample that is used to freeze this model. It’s useful for debug and visualization. It could be left unset if sample is not used when freezing the model.
It’s supposed to be a dict which is previously known as architecture dict (however it can sometimes contain information about evaluator as well).
Subclasses should set this attribute in
freeze()if they want to use it. They may also set a sample different from what they received infreeze()if it’s intended.
- status: ModelStatus¶
The status of the model space / model.
- class nni.nas.space.Graph(model, graph_id, name=None, _internal=False)[source]¶
Graph topology.
This class simply represents the topology, with no semantic meaning. All other information like metric, non-graph functions, mutation history, etc should go to
GraphModelSpace.Each graph belongs to and only belongs to one
GraphModelSpace.- model¶
The model containing (and owning) this graph.
- id¶
Unique ID in the model. If two models have graphs of identical ID, they are semantically the same graph. Typically this means one graph is mutated from another, or they are both mutated from one ancestor.
- name¶
Mnemonic name of this graph. It should have an one-to-one mapping with ID.
- input_names¶
Optional mnemonic names of input parameters.
- output_names¶
Optional mnemonic names of output values.
- input_node¶
Incoming node.
- output_node¶
Output node.
Hidden nodes
- nodes¶
All input/output/hidden nodes.
- edges¶
Edges.
- python_name¶
The name of torch.nn.Module, should have one-to-one mapping with items in python model.
- fork()[source]¶
Fork the model and returns corresponding graph in new model. This shortcut might be helpful because many algorithms only cares about “stem” subgraph instead of whole model.
- get_node_by_id(node_id)[source]¶
Returns the node which has specified name; or returns None if no node has this name.
- get_node_by_name(name)[source]¶
Returns the node which has specified name; or returns None if no node has this name.
- class nni.nas.space.GraphModelSpace(*, _internal=False)[source]¶
Represents a neural network model space with graph. Previously known as
GraphModelSpace.During mutation, one
GraphModelSpaceobject is created for each trainable snapshot. For example, consider a mutator that insert a node at an edge for each iteration. In one iteration, the mutator invokes 4 primitives: add node, remove edge, add edge to head, add edge to tail. These 4 primitives operates in oneGraphModelSpaceobject. When they are all done the model will be set to “frozen” (trainable) status and be submitted to execution engine. And then a new iteration starts, and a newGraphModelSpaceobject is created by forking last model.- status¶
See
ModelStatus.
- root_graph¶
The outermost graph which usually takes dataset as input and feeds output to loss function.
- graphs¶
All graphs (subgraphs) in this model.
- evaluator¶
GraphModelSpace evaluator
- mutators¶
List of mutators that are applied to this model.
- metrics¶
Intermediate as well as final metrics.
- fork()[source]¶
Create a new model which has same topology, names, and IDs to current one.
Can only be invoked on a frozen model. The new model will be in Mutating state.
This API is used in mutator base class.
- freeze(sample)[source]¶
Freeze the model by applying the sample to mutators.
Can only be invoked on a mutating model. The new model will be in Frozen state.
This API is used in mutator base class.
- get_node_by_name(node_name)[source]¶
Traverse all the nodes to find the matched node with the given name.
- get_node_by_python_name(python_name)[source]¶
Traverse all the nodes to find the matched node with the given python_name.
- get_nodes_by_label(label)[source]¶
Traverse all the nodes to find the matched node(s) with the given label. There could be multiple nodes with the same label. Name space name can uniquely identify a graph or node.
NOTE: the implementation does not support the class abstraction
- get_nodes_by_type(type_name)[source]¶
Traverse all the nodes to find the matched node(s) with the given type.
- class nni.nas.space.Metrics(strict=True)[source]¶
Data structure that manages the metric data (e.g., loss, accuracy, etc.).
NOTE: Multiple metrics and minimized metrics are not supported in the current iteration.
- Parameters:
strict (bool) – Whether to convert the metrics into a float. If
true, only float metrics or dict with “default” are accepted.
- class nni.nas.space.ModelStatus(value)[source]¶
The status of model space.
A model space is created in Initialized status. When the model space starts to mutate and is becoming a single model, the status will be set to Mutating. As the model space will share the same class with the mutated single model, the status flag is a useful indication for the difference between the two.
When the mutation is done and the model get ready to train, its status becomes Frozen. Only Frozen models can be submitted to execution engine for training. When training started, the model’s status becomes Training. If training is successfully ended, model’s metric attribute get set and its status becomes Trained. If training failed, the status becomes Failed.
- class nni.nas.space.Mutation(mutator, samples, from_, to)[source]¶
An execution of mutation, which consists of four parts: a mutator, a list of decisions (choices), the model that it comes from, and the model that it becomes.
In general cases, the mutation logs are not reliable and should not be replayed as the mutators can be arbitrarily complex. However, for inline mutations, the labels correspond to mutator labels here, this can be useful for metadata visualization and python execution mode.
- mutator¶
Mutator.
- samples¶
Decisions/choices.
- from_¶
Model that is comes from.
- to¶
Model that it becomes.
- class nni.nas.space.MutationSampler[source]¶
Handles
Mutator.choice()calls.Choice is the only supported type for mutator.
- class nni.nas.space.Mutator(*, sampler=None, label=None)[source]¶
Mutates graphs in model to generate new model.
By default, mutator simplifies to a single-value dict with its own label as key, and itself as value. At freeze, the strategy should provide a
MutationSamplerin the dict. This is because the freezing of mutator is dynamic (i.e., requires a variational number of random numbers, dynamic ranges for each random number), and theMutationSamplerhere can be considered as some random number generator to produce a random sequence based on the asks inMutator.mutate().On the other hand, a subclass mutator should implement
Mutator.mutate(), which callsMutator.choice()inside, andMutator.choice()invokes the bounded sampler to “random” a choice.The label of the mutator in most cases is the label of the nodes on which the mutator is applied to.
I imagine that mutating any model space (other than graph) might be useful, but we would postpone the support to when we actually need it.
- apply(model)[source]¶
Apply this mutator on a model. The model will be copied before mutation and the original model will not be modified.
- Return type:
The mutated model.
- bind_model(model)[source]¶
Mutators need a model, based on which they generate new models. This context manager binds a model to the mutator, and unbinds it after the context.
Examples
>>> with mutator.bind_model(model): ... mutator.simplify()
- bind_sampler(sampler)[source]¶
Set the sampler which will handle
Mutator.choice()calls.
- freeze(sample)[source]¶
When freezing a mutator, we need a model to mutate on, as well as a sampler to generate choices.
As how many times the mutator is applied on the model is often variational, a sample with fixed length will not work. The dict values in
sampleshould be a sampler inheritingMutationSampler. But there are also cases wheresimplify()converts the mutation process into some fixed operations (e.g., inStationaryMutator). In this case, sub-class should handle the freeze logic on their own.Mutator.freeze()needs to be called in abind_modelcontext.
- leaf_mutables(is_leaf)[source]¶
By default, treat self as a whole labeled mutable in the format dict.
Sub-class can override this to dry run the mutation upon the model and return the mutated model for the followed-up dry run.
See also
- class nni.nas.space.MutatorSequence(mutators)[source]¶
Apply a series of mutators on our model, sequentially.
This could be generalized to a DAG indicating the dependencies between mutators, but we don’t have a use case for that yet.
- class nni.nas.space.Node(graph, node_id, name, operation, _internal=False)[source]¶
An operation or an opaque subgraph inside a graph.
Each node belongs to and only belongs to one
Graph. Nodes should never be created with constructor. UseGraph.add_node()instead.The node itself is for topology only. Information of tensor calculation should all go inside
operationattribute.TODO: parameter of subgraph (cell) It’s easy to assign parameters on cell node, but it’s hard to “use” them. We need to design a way to reference stored cell parameters in inner node operations. e.g.
self.fc = Linear(self.units)<- how to expressself.unitsin IR?- graph¶
The graph containing this node.
- id¶
Unique ID in the model. If two models have nodes with same ID, they are semantically the same node.
- name¶
Mnemonic name. It should have an one-to-one mapping with ID.
- python_name¶
The name of torch.nn.Module, should have one-to-one mapping with items in python model.
- label¶
Optional. If two nodes have the same label, they are considered same by the mutator.
- operation¶
Operation.
- cell¶
Read only shortcut to get the referenced subgraph. If this node is not a subgraph (is a primitive operation), accessing
cellwill raise an error.
- predecessors¶
Predecessor nodes of this node in the graph. This is an optional mutation helper.
- successors¶
Successor nodes of this node in the graph. This is an optional mutation helper.
- incoming_edges¶
Incoming edges of this node in the graph. This is an optional mutation helper.
- outgoing_edges¶
Outgoing edges of this node in the graph. This is an optional mutation helper.
- class nni.nas.space.RawFormatModelSpace(model_space, evaluator)[source]¶
Model space that keeps the original model and does no conversion of model format (in contrast to
SimplifiedModelSpaceorGraphModelSpace).It’s possible that strategies directly operate on this format of model space, but it will be very slow (since dealing with deep learning models directly) and inflexible.
Therefore, this is almost only useful when strategies need to fuse the model space and evaluator, which requires source-code-level access to those two components. One typical use case is one-shot strategy.
In the current version,
RawFormatModelSpacecan’t be serialized and sent to remote machines.Examples
A simple example of using
RawFormatModelSpaceis as follows:from nni.nas.nn.pytorch import ModelSpace class MyModelSpace(ModelSpace): ... evaluator = FunctionEvaluator(evaluate_fn, learning_rate=nni.choice('lr', [0.1, 1.0])) model_space = RawFormatModelSpace(MyModelSpace(), evaluator)
The space can then be simplified and freezed:
frozen_model = model_space.freeze({'layer1': 0, 'lr': 0.1})
The frozen model can be instantiated and executed:
model = frozen_model.executable_model() evaluator.evaluate(model)
- class nni.nas.space.SimplifiedModelSpace(model, mutables, evaluator)[source]¶
Model space that is simplified (see
simplify()), and only keeps the key information.With
SimplifiedModelSpace, all details inside the model will be removed, which means, the weights, attributes, inplace modifications of the model will all be lost. Only the simplified mutables and necessary init arguments to recover the model for execution will be kept.The
freeze()method does nothing but remembers the sample. When the model is actually executed for real (i.e., when :meth;`executable_model` is called), the model will be recreated from scratch, and the sample will be applied to the model. To be specific, it will create the model with traced symbols and arguments, but under amodel_context(). The context can be detected viacurrent_model(). It’s the responsibility of the model space to check whether the context is available, and create a frozen model directly if it is (note thatfreezeandcontainsmethod of model space is never used).MutableModuleis an example which has already implemented this logic.
- class nni.nas.space.StationaryMutator(*, sampler=None, label=None)[source]¶
A mutator that can be dry run.
StationaryMutatorinvokeStationaryMutator.dry_runto predict choice candidates, such that the mutator simplifies to some static choices within simplify(). This could be convenient to certain algorithms which do not want to handle dynamic samplers.- dry_run(model)[source]¶
Dry run mutator on a model to collect choice candidates.
If you invoke this method multiple times on same or different models, it may or may not return identical results, depending on how the subclass implements Mutator.mutate().
Recommended to be used in
simplify()if the mutator is static.
- leaf_mutables(is_leaf)[source]¶
Simplify this mutator to a number of static choices. Invokes
StationaryMutator.dry_run().Must be wrapped in a
bind_modelcontext.
- random(memo=None, random_state=None)[source]¶
Use
nni.mutable.Mutable.random()to generate a random sample.
- nni.nas.space.current_model()[source]¶
Get the current model sample in
model_context().The sample is supposed to be the same as
nni.nas.space.ExecutableModelSpace.sample.This method is only valid when called inside
model_context(). By default, only the execution ofSimplifiedModelSpacewill set the context, so thatcurrent_model()is meaningful within the re-instantiation of the model.- Returns:
Model sample (i.e., architecture dict) before freezing, produced by strategy.
If not called inside
model_context(), returns None.
- Return type:
Dict[str, Any] | None
- nni.nas.space.model_context(sample)[source]¶
Get a context stack of the current model sample (i.e., architecture dict).
This should be used together with
current_model().model_context()is read-only, and should not be used to modify the architecture dict.
Execution engine¶
- class nni.nas.execution.ExecutionEngine[source]¶
The abstract interface of execution engine.
Execution engine is responsible for executing the submitted models. The engine has the freedom to choose the execution environment. For example, whether to execute it instantly in the current process, or send it to NNI training service (e.g., local / remote). It may also optimize the workloads with techniques like CSE, or even doing benchmark queries.
Note that some engines might reply on certain model space formats. For example, some engines might require the model space to be a graph, to do certain optimizations.
Every subclass of class:ExecutableModelSpace has its general logic (i.e., code) of execution defined in its class. But the interpretation of the logic depends on the engine itself.
In synchronized use case, the strategy will have a loop to call submit_models and wait_models repeatedly, and will receive metrics from ExecutableModelSpace attributes. Execution engine could assume that strategy will only submit graph when there are available resources (for now).
In asynchronized use case, the strategy will register a listener to receive events, while still using submit_models to train.
There might be some util functions benefit all optimizing methods, but non-mandatory utils should not be covered in abstract interface.
- budget_available()[source]¶
Return whether the engine still has available budget.
Budget could be defined by the number of models, total duration, or energy consumption, etc.
If the engine has already exhausted the budget, it will not accept any new models.
NOTE: NNI has no definition of budget yet. Therefore this method only returns true or false. In future, we might change it to a concrete budget.
- default_callback(event)[source]¶
Default callback that is called when a model has a new metric, or a new status.
This callback is called after all callbacks registered by the user of this engine, if it’s not canceled.
The callback implements the most typical behavior of an event:
Update the metrics of the model if the event is a metric event.
Update the status of the model if the event is a status event.
- dispatch_model_event(event: ModelEventType, **kwargs: Any) None[source]¶
- dispatch_model_event(event: str, **kwargs: Any) None
- dispatch_model_event(event: ModelEvent) None
Dispatch a model event to all callbacks. Invoke
default_callback()at the end. This is a utility method for subclass ofExecutionEngineto dispatch (emit) events.If the engine intends to change the model status / metrics, and also notifies the listeners, they are supposed to construct a model event and call
dispatch_model_event(), rather than changing the status of metrics of the model directly. Only in this way, the listeners can properly receive the update, and even intercept the update before they actually take effect.The behavior of
default_callback()is defined by whoever “dispatches” the event (although it has a default implementation).
- idle_worker_available()[source]¶
Return the number of idle workers. That is, the recommended number of models to submit currently.
Strategy can respect / ignore the number. If strategy chooses to ignore, the engine doesn’t guarantee anything about the newly-submitted model.
NOTE: The return value was originally designed to be a list of
WorkerInfoobjects. If no details are available, this may returns a list of “empty” objects, reporting the number of idle workers. However,WorkerInfois almost never used in practice. So we removed it for now to simplify the type-checking.
- list_models(status=None)[source]¶
Get all models submitted.
If status is presented, only return models with the given status.
Execution engine should store a copy of models that have been submitted and return a list of copies in this method.
- load_state_dict(state_dict)[source]¶
Load the state of the engine.
Symmetric to
state_dict().
- register_model_event_callback(event_type, callback)[source]¶
Register a callback to receive model event.
- Parameters:
event_type (ModelEventType) – The type of event that is to listen.
callback (Callable[[...], None]) – The callback to receive the event. It receives a
ModelEventobject, and is expected to return nothing.
- shutdown()[source]¶
Stop the engine.
The engine will not accept new models, or handle callbacks after being shutdown. Anything after
shutdown()is called is considered undefined behavior.Since engine is ephemeral, there is no such thing as
restart. Creating another engine and load the state dict is encouraged instead.
- submit_models(*models)[source]¶
Submit models to NNI.
This method is supposed to call something like nni.Advisor.create_trial_job(graph_data).
- unregister_model_event_callback(event_type, callback)[source]¶
Unregister a callback.
- Parameters:
event_type (ModelEventType) – The type of event that is to listen.
callback (Callable[[...], None]) – The callback to receive the event. The event must have been registered before.
- class nni.nas.execution.FinalMetricEvent(model, metric)[source]¶
Event of a model update with final metric.
Currently the metric is raw, and wasn’t canonicalized. But it’s subject to change in next iterations.
- class nni.nas.execution.IntermediateMetricEvent(model, metric)[source]¶
Event of a model update with intermediate metric.
- class nni.nas.execution.Middleware(engine=None)[source]¶
A middleware that wraps another execution engine. It can be used to transform the submitted models before passing to the underlying engine.
Middlewares sits between a strategy and a engine. There could be multiple middlewares chained. Some middlewares logically belong to the strategy side, for example model filters and early stopper. Others logically belong to the engine side, for example CSE and benchmarking. This class is designed mainly for the engine side. Strategy side should inherit another dedicated superclass.
Implementing a middleware is similar to implementing an engine, but with the option of leveraging the ability of the underlying wrapped engine. Apart from the methods that would otherwise raise NotImplementedError if not implemented, we recommend override
set_engine()andregister_model_event_callback(). Inset_engine(), the middleware registers some callbacks by itself on the underlying engine, while inregister_model_event_callback(), the middleware decides what to do with the callbacks from the outside. There are basically two approaches to handle the callbacks:Register the callbacks directly on the underlying engine. Since callbacks in
set_engine()are registered before the callbacks from the outside, they can intercept the events and manipulates/stops them when needed.Keep the callbacks to itself. Register callbacks written by the middleware itself to the underlying engine, which creates brand new events and uses
dispatch_model_event()to invoke the callbacks from the outside.
Some other (hacky) approaches might not be possible (e.g., wrap the callbacks with a closure). But they are not recommended.
Middleware should be responsible for unregistering the callbacks at
shutdown().- Parameters:
engine (ExecutionEngine | None) – The underlying execution engine.
- property engine: ExecutionEngine¶
The underlying execution engine (or another middleware).
- set_engine(engine)[source]¶
Override this to do some initialization, e.g., register some callbacks.
Engine can’t be “unset” once set, because middlewares can be only binded once. To unregister the callbacks, override
shutdown().- Parameters:
engine (ExecutionEngine) – The underlying execution engine.
- class nni.nas.execution.ModelEvent(model)[source]¶
Event of a model update.
- class nni.nas.execution.SequentialExecutionEngine(max_model_count=None, max_duration=None, continue_on_failure=False)[source]¶
The execution engine will run every model in the current process. If multiple models have been submitted, they will be queued and run sequentially.
Keyboard interrupt will terminate the currently running model and raise to let the main process know.
- class nni.nas.execution.TrainingEndEvent(model, status)[source]¶
Event of a model update with training end.
- class nni.nas.execution.TrainingServiceExecutionEngine(nodejs_binding, fetch_intermediates=True)[source]¶
The execution engine will submit every model onto training service.
Resource management is implemented in this class.
This engine doesn’t include any optimization across graphs.
NOTE: Due to the design of nni.experiment, the execution engine resorts to NasExperiment to submit trials as well as waiting for results. This is not ideal, because this engine might be one of the very few engines which need the training service. Ideally, the training service should be a part of the execution engine, not the experiment.
Ideally, this class should not have any states. Its save and load methods should be empty.
- Parameters:
nodejs_binding (NasExperiment) – The nodejs binding of the experiment.
fetch_intermediates (bool) – Whether to fetch intermediate results from the training service when list models. Setting it to false for large-scale experiments can improve performance.
- budget_available()[source]¶
Infer the budget from resources.
This should have a dedicated implementation on the nodejs side in the future.
- idle_worker_available()[source]¶
Return the number of available resources.
The resource is maintained by the engine itself. It should be fetched from nodejs side directly in future.
- list_models(status=None)[source]¶
Retrieve models previously submitted.
To support a large-scale experiments with thousands of trials, this method will retrieve the models from the nodejs binding (i.e., from the database). The model instances will be re-created on the fly based on the data from database. Although they are the same models semantically, they might not be the same instances. Exceptions are those still used by the strategy. Their weak references are kept in the engine and thus the exact same instances are returned.
- Parameters:
status (ModelStatus | None) – The status of the models to be retrieved. If None, all models will be retrieved.
include_intermediates – Whether to include intermediate models.
- submit_models(*models)[source]¶
Submit models to training service.
See also
nni.nas.ExecutionEngine.submit_models
- wait_models(*models)[source]¶
Wait models to finish training.
If argument models is empty, wait for all models to finish. Using the experiment status as an indicator of all models’ status, which is more efficient.
For the models to receive status changes, the models must be the exact same instances as the ones submitted. Dumping and reloading the models, or retrieving the unsaved models from
list_models()won’t work.
Cross-graph optimization¶
- class nni.nas.execution.cgo.CrossGraphOptimization(remote_config, max_concurrency=None, batch_waiting_time=60)[source]¶
The execution engine middleware of Cross-Graph Optimization (CGO). It’s a technique that merges multiple models into one model for training speedup. See Retiarii paper for details.
Currently,
CrossGraphOptimizationis only a prototype. It’s not fully tested, and also, comes with a bunch of constraints on the model space and evaluator:The models must be in the format of
GraphModelSpace.The evaluator has to be a
Lightningevaluator.The
lightning_moduleargument of the evaluator must be an instance ofMultiModelSupervisedLearningModule.The
trainerargument of the evaluator must be an instance ofMultiModelTrainer.
There are also a number of limitations:
CGO doesn’t support stop and resume a checkpoint.
Only remote training service is supported.
All model history are stored in memory. The experiment might not scale well.
- Parameters:
remote_config (RemoteConfig) – The remote training service config.
max_concurrency (int | None) – The maximum number of trials to run concurrently.
batch_waiting_time (int) – Seconds to wait for each batch of trial submission. The trials within one batch could apply cross-graph optimization.
- class nni.nas.execution.cgo.MultiModelLightningModule(criterion, metric, n_models=None)[source]¶
The lightning module for a merged “multi-model”.
The output of the multi-model is expected to be a tuple of tensors. The tensors will be each passed to a criterion and a metric. The loss will be added up for back propagation, and the metrics will be logged.
The reported metric will be a list of metrics, one for each model.
- Parameters:
criterion (nn.Module) – Loss function.
metric (Metric) – Metric function.
n_models (int | None) – Number of models in the multi-model.
- class nni.nas.execution.cgo.MultiModelTrainer(*args, **kwargs)[source]¶
Trainer for cross-graph optimization.
- Parameters:
use_cgo (bool) – Whether cross-graph optimization (CGO) is used. If it is True, CGO will manage device placement. Any device placement from pytorch lightning will be bypassed. default: False
trainer_kwargs – Optional keyword arguments passed to trainer. See Lightning documentation for details.
NAS Benchmarks¶
- class nni.nas.benchmark.BenchmarkEvaluator[source]¶
A special kind of evaluator that does not run real training, but queries a database.
- classmethod default_space()[source]¶
Return the default search space benchmarked by this evaluator.
Subclass should override this.
- evaluate(sample)[source]¶
evaluate()receives a sample and returns a float score. It also reports intermediate and final results through NNI trial API.Necessary format conversion and database query should be done in this method.
It is the main interface of this class. Subclass should override this.
- validate_space(space)[source]¶
Validate the search space. Raise exception if invalid. Returns the validated space.
By default, it will cross-check with the
default_space(), and return the default space. Differences in common scope names will be ignored.I think the default implementation should work for most cases. But subclass can still override this method for looser or tighter validation.
- class nni.nas.benchmark.BenchmarkModelSpace(model_space: BenchmarkEvaluator)[source]¶
- class nni.nas.benchmark.BenchmarkModelSpace(model_space: BaseModelSpace)
- class nni.nas.benchmark.BenchmarkModelSpace(model_space: None, evaluator: BenchmarkEvaluator)
Model space that is specialized for benchmarking.
We recommend using this model space for benchmarking, for its validation and efficiency.
- Parameters:
model_space (BaseModelSpace | BenchmarkEvaluator | None) – If not provided, it will be set to the default model space of the evaluator.
evaluator (Evaluator | None) – Evaluator that will be used to benchmark the space.
Examples
Can be either:
BenchmarkModelSpace(evaluator)
or:
BenchmarkModelSpace(pytorch_model_space, evaluator)
In the case where the model space is provided, it will be validated by the evaluator and must be a match.
- class nni.nas.benchmark.NasBench101Benchmark(num_epochs=108, metric='valid_acc', include_intermediates=False)[source]¶
Benchmark evaluator for NAS-Bench-101.
- Parameters:
num_epochs (int) – Queried
num_epochs.metric (str) – Queried metric.
include_intermediates (bool) – Whether to report intermediate results.
- class nni.nas.benchmark.NasBench201Benchmark(num_epochs=200, dataset='cifar100', metric='valid_acc', include_intermediates=False)[source]¶
Benchmark evaluator for NAS-Bench-201.
- Parameters:
num_epochs (int) – Queried
num_epochs.dataset (str) – Queried
dataset.metric (str) – Queried metric.
include_intermediates (bool) – Whether to report intermediate results.
- class nni.nas.benchmark.SlimBenchmarkSpace(mutables=None, **mutable_kwargs)[source]¶
Example model space without deep learning frameworks.
When constructing this, the dict should’ve been already simplified and validated.
It could look like:
{ 'layer1': nni.choice('layer1', ['a', 'b', 'c']), 'layer2': nni.choice('layer2', ['d', 'e', 'f']), }
- nni.nas.benchmark.download_benchmark(benchmark, progress=True)[source]¶
Download a converted benchmark.
- Parameters:
benchmark (str) – Benchmark name like nasbench201.
- nni.nas.benchmark.load_benchmark(benchmark)[source]¶
Load a benchmark as a database.
Parmaeters¶
- benchmarkstr
Benchmark name like nasbench201.
NAS-Bench-101¶
- class nni.nas.benchmark.nasbench101.Nb101IntermediateStats(*args, **kwargs)[source]¶
Intermediate statistics for NAS-Bench-101.
- trial¶
The exact trial where the intermediate result is produced.
- Type:
- current_epoch¶
Elapsed epochs when evaluation is done.
- Type:
int
- train_acc¶
Intermediate accuracy on training data, ranging from 0 to 100.
- Type:
float
- valid_acc¶
Intermediate accuracy on validation data, ranging from 0 to 100.
- Type:
float
- test_acc¶
Intermediate accuracy on test data, ranging from 0 to 100.
- Type:
float
- training_time¶
Time elapsed in seconds.
- Type:
float
- class nni.nas.benchmark.nasbench101.Nb101TrialConfig(*args, **kwargs)[source]¶
Trial config for NAS-Bench-101.
- arch¶
A dict with keys
op1,op2, … andinput1,input2, … Vertices are enumerate from 0. Since node 0 is input node, it is skipped in this dict. Eachopis one ofnni.nas.benchmark.nasbench101.CONV3X3_BN_RELU,nni.nas.benchmark.nasbench101.CONV1X1_BN_RELU, andnni.nas.benchmark.nasbench101.MAXPOOL3X3. Eachinputis a list of previous nodes. For exampleinput5can be[0, 1, 3].- Type:
dict
- num_vertices¶
Number of vertices (nodes) in one cell. Should be less than or equal to 7 in default setup.
- Type:
int
- hash¶
Graph-invariant MD5 string for this architecture.
- Type:
str
- num_epochs¶
Number of epochs planned for this trial. Should be one of 4, 12, 36, 108 in default setup.
- Type:
int
- class nni.nas.benchmark.nasbench101.Nb101TrialStats(*args, **kwargs)[source]¶
Computation statistics for NAS-Bench-101. Each corresponds to one trial. Each config has multiple trials with different random seeds, but unfortunately seed for each trial is unavailable. NAS-Bench-101 trains and evaluates on CIFAR-10 by default. The original training set is divided into 40k training images and 10k validation images, and the original validation set is used for test only.
- config¶
Setup for this trial data.
- Type:
- train_acc¶
Final accuracy on training data, ranging from 0 to 100.
- Type:
float
- valid_acc¶
Final accuracy on validation data, ranging from 0 to 100.
- Type:
float
- test_acc¶
Final accuracy on test data, ranging from 0 to 100.
- Type:
float
- parameters¶
Number of trainable parameters in million.
- Type:
float
- training_time¶
Duration of training in seconds.
- Type:
float
- nni.nas.benchmark.nasbench101.query_nb101_trial_stats(arch, num_epochs, isomorphism=True, reduction=None, include_intermediates=False)[source]¶
Query trial stats of NAS-Bench-101 given conditions.
- Parameters:
arch (dict or None) – If a dict, it is in the format that is described in
nni.nas.benchmark.nasbench101.Nb101TrialConfig. Only trial stats matched will be returned. If none, all architectures in the database will be matched.num_epochs (int or None) – If int, matching results will be returned. Otherwise a wildcard.
isomorphism (boolean) – Whether to match essentially-same architecture, i.e., architecture with the same graph-invariant hash value.
reduction (str or None) – If ‘none’ or None, all trial stats will be returned directly. If ‘mean’, fields in trial stats will be averaged given the same trial config.
include_intermediates (boolean) – If true, intermediate results will be returned.
- Returns:
A generator of
nni.nas.benchmark.nasbench101.Nb101TrialStatsobjects, where each of them has been converted into a dict.- Return type:
generator of dict
NAS-Bench-201¶
- class nni.nas.benchmark.nasbench201.Nb201IntermediateStats(*args, **kwargs)[source]¶
Intermediate statistics for NAS-Bench-201.
- trial¶
Corresponding trial.
- Type:
- current_epoch¶
Elapsed epochs.
- Type:
int
- train_acc¶
Current accuracy on training data, ranging from 0 to 100.
- Type:
float
- valid_acc¶
Current accuracy on validation data, ranging from 0 to 100.
- Type:
float
- test_acc¶
Current accuracy on test data, ranging from 0 to 100.
- Type:
float
- ori_test_acc¶
Test accuracy on original validation set (10k for CIFAR and 12k for Imagenet16-120), ranging from 0 to 100.
- Type:
float
- train_loss¶
Current cross entropy loss on training data.
- Type:
float or None
- valid_loss¶
Current cross entropy loss on validation data.
- Type:
float or None
- test_loss¶
Current cross entropy loss on test data.
- Type:
float or None
- ori_test_loss¶
Current cross entropy loss on original validation set.
- Type:
float or None
- class nni.nas.benchmark.nasbench201.Nb201TrialConfig(*args, **kwargs)[source]¶
Trial config for NAS-Bench-201.
- arch¶
A dict with keys
0_1,0_2,0_3,1_2,1_3,2_3, each of which is an operator chosen fromnni.nas.benchmark.nasbench201.NONE,nni.nas.benchmark.nasbench201.SKIP_CONNECT,nni.nas.benchmark.nasbench201.CONV_1X1,nni.nas.benchmark.nasbench201.CONV_3X3andnni.nas.benchmark.nasbench201.AVG_POOL_3X3.- Type:
dict
- num_epochs¶
Number of epochs planned for this trial. Should be one of 12 and 200.
- Type:
int
- num_channels¶
Number of channels for initial convolution. 16 by default.
- Type:
int
- num_cells¶
Number of cells per stage. 5 by default.
- Type:
int
- dataset¶
Dataset used for training and evaluation. NAS-Bench-201 provides the following 4 options:
cifar10-valid(training data is splited into 25k for training and 25k for validation, validation data is used for test),cifar10(training data is used in training, validation data is splited into 5k for validation and 5k for testing),cifar100(same protocol ascifar10), andimagenet16-120(a subset of 120 classes in ImageNet, downscaled to 16x16, using training data for training, 6k images from validation set for validation and the other 6k for testing).- Type:
str
- class nni.nas.benchmark.nasbench201.Nb201TrialStats(*args, **kwargs)[source]¶
Computation statistics for NAS-Bench-201. Each corresponds to one trial.
- config¶
Setup for this trial data.
- Type:
- seed¶
Random seed selected, for reproduction.
- Type:
int
- train_acc¶
Final accuracy on training data, ranging from 0 to 100.
- Type:
float
- valid_acc¶
Final accuracy on validation data, ranging from 0 to 100.
- Type:
float
- test_acc¶
Final accuracy on test data, ranging from 0 to 100.
- Type:
float
- ori_test_acc¶
Test accuracy on original validation set (10k for CIFAR and 12k for Imagenet16-120), ranging from 0 to 100.
- Type:
float
- train_loss¶
Final cross entropy loss on training data. Note that loss could be NaN, in which case this attributed will be None.
- Type:
float or None
- valid_loss¶
Final cross entropy loss on validation data.
- Type:
float or None
- test_loss¶
Final cross entropy loss on test data.
- Type:
float or None
- ori_test_loss¶
Final cross entropy loss on original validation set.
- Type:
float or None
- parameters¶
Number of trainable parameters in million.
- Type:
float
- latency¶
Latency in seconds.
- Type:
float
- flops¶
FLOPs in million.
- Type:
float
- training_time¶
Duration of training in seconds.
- Type:
float
- valid_evaluation_time¶
Time elapsed to evaluate on validation set.
- Type:
float
- test_evaluation_time¶
Time elapsed to evaluate on test set.
- Type:
float
- ori_test_evaluation_time¶
Time elapsed to evaluate on original test set.
- Type:
float
- nni.nas.benchmark.nasbench201.query_nb201_trial_stats(arch, num_epochs, dataset, reduction=None, include_intermediates=False)[source]¶
Query trial stats of NAS-Bench-201 given conditions.
- Parameters:
arch (dict or None) – If a dict, it is in the format that is described in
nni.nas.benchmark.nasbench201.Nb201TrialConfig. Only trial stats matched will be returned. If none, all architectures in the database will be matched.num_epochs (int or None) – If int, matching results will be returned. Otherwise a wildcard.
dataset (str or None) – If specified, can be one of the dataset available in
nni.nas.benchmark.nasbench201.Nb201TrialConfig. Otherwise a wildcard.reduction (str or None) – If ‘none’ or None, all trial stats will be returned directly. If ‘mean’, fields in trial stats will be averaged given the same trial config.
include_intermediates (boolean) – If true, intermediate results will be returned.
- Returns:
A generator of
nni.nas.benchmark.nasbench201.Nb201TrialStatsobjects, where each of them has been converted into a dict.- Return type:
generator of dict
NDS¶
- class nni.nas.benchmark.nds.NdsIntermediateStats(*args, **kwargs)[source]¶
Intermediate statistics for NDS.
- trial¶
Corresponding trial.
- Type:
- current_epoch¶
Elapsed epochs.
- Type:
int
- train_loss¶
Current cross entropy loss on training data. Can be NaN (None).
- Type:
float or None
- train_acc¶
Current accuracy on training data, ranging from 0 to 100.
- Type:
float
- test_acc¶
Current accuracy on test data, ranging from 0 to 100.
- Type:
float
- class nni.nas.benchmark.nds.NdsTrialConfig(*args, **kwargs)[source]¶
Trial config for NDS.
- model_family¶
Could be
nas_cell,residual_bottleneck,residual_basicorvanilla.- Type:
str
- model_spec¶
If
model_familyisnas_cell, it containsnum_nodes_normal,num_nodes_reduce,depth,width,auxanddrop_prob. Ifmodel_familyisresidual_bottleneck, it containsbot_muls,ds(depths),num_gs(number of groups) andss(strides). Ifmodel_familyisresidual_basicorvanilla, it containsds,ssandws.- Type:
dict
- cell_spec¶
If
model_familyis notnas_cellit will be an empty dict. Otherwise, it specifies<normal/reduce>_<i>_<op/input>_<x/y>, where i ranges from 0 tonum_nodes_<normal/reduce> - 1. If it is anop, the value is chosen from the constants specified previously likenni.nas.benchmark.nds.CONV_1X1. If it is i’sinput, the value range from 0 toi + 1, asnas_celluses previous two nodes as inputs, and node 0 is actually the second node. Refer to NASNet paper for details. Finally, another two key-value pairsnormal_concatandreduce_concatspecify which nodes are eventually concatenated into output.- Type:
dict
- dataset¶
Dataset used. Could be
cifar10orimagenet.- Type:
str
- generator¶
Can be one of
randomwhich generates configurations at random, while keeping learning rate and weight decay fixed,fix_w_dwhich further keepswidthanddepthfixed, only applicable fornas_cell.tune_lr_wdwhich further tunes learning rate and weight decay.- Type:
str
- proposer¶
Paper who has proposed the distribution for random sampling. Available proposers include
nasnet,darts,enas,pnas,amoeba,vanilla,resnext-a,resnext-b,resnet,resnet-b(ResNet with bottleneck). See NDS paper for details.- Type:
str
- base_lr¶
Initial learning rate.
- Type:
float
- weight_decay¶
L2 weight decay applied on weights.
- Type:
float
- num_epochs¶
Number of epochs scheduled, during which learning rate will decay to 0 following cosine annealing.
- Type:
int
- class nni.nas.benchmark.nds.NdsTrialStats(*args, **kwargs)[source]¶
Computation statistics for NDS. Each corresponds to one trial.
- config¶
Corresponding config for trial.
- Type:
- seed¶
Random seed selected, for reproduction.
- Type:
int
- final_train_acc¶
Final accuracy on training data, ranging from 0 to 100.
- Type:
float
- final_train_loss¶
Final cross entropy loss on training data. Could be NaN (None).
- Type:
float or None
- final_test_acc¶
Final accuracy on test data, ranging from 0 to 100.
- Type:
float
- best_train_acc¶
Best accuracy on training data, ranging from 0 to 100.
- Type:
float
- best_train_loss¶
Best cross entropy loss on training data. Could be NaN (None).
- Type:
float or None
- best_test_acc¶
Best accuracy on test data, ranging from 0 to 100.
- Type:
float
- parameters¶
Number of trainable parameters in million.
- Type:
float
- flops¶
FLOPs in million.
- Type:
float
- iter_time¶
Seconds elapsed for each iteration.
- Type:
float
- nni.nas.benchmark.nds.query_nds_trial_stats(model_family, proposer, generator, model_spec, cell_spec, dataset, num_epochs=None, reduction=None, include_intermediates=False)[source]¶
Query trial stats of NDS given conditions.
- Parameters:
model_family (str or None) – If str, can be one of the model families available in
nni.nas.benchmark.nds.NdsTrialConfig. Otherwise a wildcard.proposer (str or None) – If str, can be one of the proposers available in
nni.nas.benchmark.nds.NdsTrialConfig. Otherwise a wildcard.generator (str or None) – If str, can be one of the generators available in
nni.nas.benchmark.nds.NdsTrialConfig. Otherwise a wildcard.model_spec (dict or None) – If specified, can be one of the model spec available in
nni.nas.benchmark.nds.NdsTrialConfig. Otherwise a wildcard.cell_spec (dict or None) – If specified, can be one of the cell spec available in
nni.nas.benchmark.nds.NdsTrialConfig. Otherwise a wildcard.dataset (str or None) – If str, can be one of the datasets available in
nni.nas.benchmark.nds.NdsTrialConfig. Otherwise a wildcard.num_epochs (float or None) – If int, matching results will be returned. Otherwise a wildcard.
reduction (str or None) – If ‘none’ or None, all trial stats will be returned directly. If ‘mean’, fields in trial stats will be averaged given the same trial config.
include_intermediates (boolean) – If true, intermediate results will be returned.
- Returns:
A generator of
nni.nas.benchmark.nds.NdsTrialStatsobjects, where each of them has been converted into a dict.- Return type:
generator of dict
Miscellaneous Utilities¶
This module does what CheckpointIO does in PyTorch-Lightning, but with a simpler implementation and a wider range of backend supports.
The default implementation is TorchSerializer, which uses torch.save() and torch.load()
to save and load the data. But it can’t be used in some cases, e.g., when torch is not installed,
or when the data requires special handling that are not supported by torch.
There are several alternatives, which can be switched via set_default_serializer().
The serializer defined in nni.common.serializer happened to be one of the alternatives.
NOTE: The file is placed in NAS experimentally. It might be merged with the global serializer in near future.
- class nni.nas.utils.serializer.JsonSerializer[source]¶
The serializer that utilizes
nni.dump()andnni.load()to save and load data.This serializer should work in cases where strategies have no complex objects in their states. Since it uses
nni.dump(), it resorts to binary format when some part of the data is not JSON-serializable.
- class nni.nas.utils.serializer.Serializer[source]¶
Save data to a file, or load data from a file.
- load(path)[source]¶
Load the data from the given path.
- Raises:
FileNotFoundError – If the file (suffixed with
suffix) is not found.
- suffix: ClassVar[str] = ''¶
All serializers should save the file with a suffix, which is used to validate the serializer type when loading data.
- class nni.nas.utils.serializer.TorchSerializer(map_location=None)[source]¶
The serializer that utilizes
torch.save()andtorch.load()to save and load data.This serializer should work in most scenarios, including cases when strategies have some tensors in their states (e.g., DRL). The downside is that it relies on torch to be installed.
- Parameters:
map_location (Any) – The
map_locationargument to be passed totorch.load().
- nni.nas.utils.serializer.get_default_serializer()[source]¶
Get the default serializer.
- Return type:
The default serializer.
- nni.nas.utils.serializer.set_default_serializer(serializer)[source]¶
Set the default serializer.
- Parameters:
serializer (Serializer) – The serializer to be used as default.