Search Space¶
Mutation Pritimives¶
LayerChoice¶
- class nni.retiarii.nn.pytorch.LayerChoice(candidates, *, prior=None, label=None, **kwargs)[source]¶
Layer choice selects one of the
candidates, then apply it on inputs and return results.It allows users to put several candidate operations (e.g., PyTorch modules), one of them is chosen in each explored model.
New in v2.2: Layer choice can be nested.
- Parameters:
candidates (list of nn.Module or OrderedDict) – A module list to be selected from.
prior (list of float) – Prior distribution used in random sampling.
label (str) – Identifier of the layer choice.
- length¶
Deprecated. Number of ops to choose from.
len(layer_choice)is recommended.- Type:
int
- names¶
Names of candidates.
- Type:
list of str
- choices¶
Deprecated. A list of all candidate modules in the layer choice module.
list(layer_choice)is recommended, which will serve the same purpose.- Type:
list of Module
Examples
# import nni.retiarii.nn.pytorch as nn # declared in `__init__` method self.layer = nn.LayerChoice([ ops.PoolBN('max', channels, 3, stride, 1), ops.SepConv(channels, channels, 3, stride, 1), nn.Identity() ]) # invoked in `forward` method out = self.layer(x)
Notes
candidatescan be a list of modules or a ordered dict of named modules, for example,self.op_choice = LayerChoice(OrderedDict([ ("conv3x3", nn.Conv2d(3, 16, 128)), ("conv5x5", nn.Conv2d(5, 16, 128)), ("conv7x7", nn.Conv2d(7, 16, 128)) ]))
Elements in layer choice can be modified or deleted. Use
del self.op_choice["conv5x5"]orself.op_choice[1] = nn.Conv3d(...). Adding more choices is not supported yet.
InputChoice¶
- class nni.retiarii.nn.pytorch.InputChoice(n_candidates, n_chosen=1, reduction='sum', *, prior=None, label=None, **kwargs)[source]¶
Input choice selects
n_choseninputs fromchoose_from(containsn_candidateskeys).It is mainly for choosing (or trying) different connections. It takes several tensors and chooses
n_chosentensors from them. When specific inputs are chosen,InputChoicewill becomeChosenInputs.Use
reductionto specify how chosen inputs are reduced into one output. A few options are:none: do nothing and return the list directly.sum: summing all the chosen inputs.mean: taking the average of all chosen inputs.concat: concatenate all chosen inputs at dimension 1.
We don’t support customizing reduction yet.
- Parameters:
n_candidates (int) – Number of inputs to choose from. It is required.
n_chosen (int) – Recommended inputs to choose. If None, mutator is instructed to select any.
reduction (str) –
mean,concat,sumornone.prior (list of float) – Prior distribution used in random sampling.
label (str) – Identifier of the input choice.
Examples
# import nni.retiarii.nn.pytorch as nn # declared in `__init__` method self.input_switch = nn.InputChoice(n_chosen=1) # invoked in `forward` method, choose one from the three out = self.input_switch([tensor1, tensor2, tensor3])
- class nni.retiarii.nn.pytorch.ChosenInputs(chosen, reduction)[source]¶
A module that chooses from a tensor list and outputs a reduced tensor. The already-chosen version of InputChoice.
When forward,
chosenwill be used to select inputs fromcandidate_inputs, andreductionwill be used to choose from those inputs to form a tensor.- chosen¶
Indices of chosen inputs.
- Type:
list of int
- reduction¶
How to reduce the inputs when multiple are selected.
- Type:
mean|concat|sum|none
ValueChoice¶
- class nni.retiarii.nn.pytorch.ValueChoice(candidates, *, prior=None, label=None)[source]¶
ValueChoice is to choose one from
candidates. The most common use cases are:Used as input arguments of
basic_unit(i.e., modules innni.retiarii.nn.pytorchand user-defined modules decorated with@basic_unit).Used as input arguments of evaluator (new in v2.7).
It can be used in parameters of operators (i.e., a sub-module of the model):
class Net(nn.Module): def __init__(self): super().__init__() self.conv = nn.Conv2d(3, nn.ValueChoice([32, 64]), kernel_size=nn.ValueChoice([3, 5, 7])) def forward(self, x): return self.conv(x)
Or evaluator (only if the evaluator is traceable, e.g.,
FunctionalEvaluator):def train_and_evaluate(model_cls, learning_rate): ... self.evaluator = FunctionalEvaluator(train_and_evaluate, learning_rate=nn.ValueChoice([1e-3, 1e-2, 1e-1]))
Value choices supports arithmetic operators, which is particularly useful when searching for a network width multiplier:
# init scale = nn.ValueChoice([1.0, 1.5, 2.0]) self.conv1 = nn.Conv2d(3, round(scale * 16)) self.conv2 = nn.Conv2d(round(scale * 16), round(scale * 64)) self.conv3 = nn.Conv2d(round(scale * 64), round(scale * 256)) # forward return self.conv3(self.conv2(self.conv1(x)))
Or when kernel size and padding are coupled so as to keep the output size constant:
# init ks = nn.ValueChoice([3, 5, 7]) self.conv = nn.Conv2d(3, 16, kernel_size=ks, padding=(ks - 1) // 2) # forward return self.conv(x)
Or when several layers are concatenated for a final layer.
# init self.linear1 = nn.Linear(3, nn.ValueChoice([1, 2, 3], label='a')) self.linear2 = nn.Linear(3, nn.ValueChoice([4, 5, 6], label='b')) self.final = nn.Linear(nn.ValueChoice([1, 2, 3], label='a') + nn.ValueChoice([4, 5, 6], label='b'), 2) # forward return self.final(torch.cat([self.linear1(x), self.linear2(x)], 1))
Some advanced operators are also provided, such as
ValueChoice.max()andValueChoice.cond().Tip
All the APIs have an optional argument called
label, mutations with the same label will share the same choice. A typical example is,self.net = nn.Sequential( nn.Linear(10, nn.ValueChoice([32, 64, 128], label='hidden_dim')), nn.Linear(nn.ValueChoice([32, 64, 128], label='hidden_dim'), 3) )
Sharing the same value choice instance has the similar effect.
class Net(nn.Module): def __init__(self): super().__init__() hidden_dim = nn.ValueChoice([128, 512]) self.fc = nn.Sequential( nn.Linear(64, hidden_dim), nn.Linear(hidden_dim, 10) )
Warning
It looks as if a specific candidate has been chosen (e.g., how it looks like when you can put
ValueChoiceas a parameter ofnn.Conv2d), but in fact it’s a syntax sugar as because the basic units and evaluators do all the underlying works. That means, you cannot assume thatValueChoicecan be used in the same way as its candidates. For example, the following usage will NOT work:self.blocks = [] for i in range(nn.ValueChoice([1, 2, 3])): self.blocks.append(Block()) # NOTE: instead you should probably write # self.blocks = nn.Repeat(Block(), (1, 3))
Another use case is to initialize the values to choose from in init and call the module in forward to get the chosen value. Usually, this is used to pass a mutable value to a functional API like
torch.xxxornn.functional.xxx`. For example,class Net(nn.Module): def __init__(self): super().__init__() self.dropout_rate = nn.ValueChoice([0., 1.]) def forward(self, x): return F.dropout(x, self.dropout_rate())
- Parameters:
candidates (list) – List of values to choose from.
prior (list of float) – Prior distribution to sample from.
label (str) – Identifier of the value choice.
- all_options()¶
Explore all possibilities of a value choice.
- static condition(pred, true, false)¶
Return
trueif the predicatepredis true elsefalse.Examples
>>> ValueChoice.condition(ValueChoice([1, 2]) > ValueChoice([0, 3]), 2, 1)
Notes
This function performs lazy evaluation. Only the expression will be recorded when the function is called. The real evaluation happens when the inner value choice has determined its final decision. If no value choice is contained in the parameter list, the evaluation will be intermediate.
- evaluate(values)¶
Evaluate the result of this group.
valuesshould in the same order ofinner_choices().
- forward()[source]¶
The forward of input choice is simply the first value of
candidates. It shouldn’t be called directly by users in most cases.
- static max(arg0, *args)¶
Returns the maximum value from a list of value choices. The usage should be similar to Python’s built-in value choices, where the parameters could be an iterable, or at least two arguments.
Notes
This function performs lazy evaluation. Only the expression will be recorded when the function is called. The real evaluation happens when the inner value choice has determined its final decision. If no value choice is contained in the parameter list, the evaluation will be intermediate.
- static min(arg0, *args)¶
Returns the minunum value from a list of value choices. The usage should be similar to Python’s built-in value choices, where the parameters could be an iterable, or at least two arguments.
Notes
This function performs lazy evaluation. Only the expression will be recorded when the function is called. The real evaluation happens when the inner value choice has determined its final decision. If no value choice is contained in the parameter list, the evaluation will be intermediate.
- static to_float(obj)¶
Convert a
ValueChoiceto a float.Notes
This function performs lazy evaluation. Only the expression will be recorded when the function is called. The real evaluation happens when the inner value choice has determined its final decision. If no value choice is contained in the parameter list, the evaluation will be intermediate.
- static to_int(obj)¶
Convert a
ValueChoiceto an integer.Notes
This function performs lazy evaluation. Only the expression will be recorded when the function is called. The real evaluation happens when the inner value choice has determined its final decision. If no value choice is contained in the parameter list, the evaluation will be intermediate.
ModelParameterChoice¶
- class nni.retiarii.nn.pytorch.ModelParameterChoice(candidates, *, prior=None, default=None, label=None)[source]¶
ModelParameterChoice chooses one hyper-parameter from
candidates.Attention
This API is internal, and does not guarantee forward-compatibility.
It’s quite similar to
ValueChoice, but unlikeValueChoice, it always returns a fixed value, even at the construction of base model.This makes it highly flexible (e.g., can be used in for-loop, if-condition, as argument of any function). For example:
self.has_auxiliary_head = ModelParameterChoice([False, True]) # this will raise error if you use `ValueChoice` if self.has_auxiliary_head is True: # or self.has_auxiliary_head self.auxiliary_head = Head() else: self.auxiliary_head = None print(type(self.has_auxiliary_head)) # <class 'bool'>
The working mechanism of
ModelParameterChoiceis that, it registers itself in themodel_wrapper, as a hyper-parameter of the model, and then returns the value specified withdefault. At base model construction, the default value will be used (as a mocked hyper-parameter). In trial, the hyper-parameter selected by strategy will be used.Although flexible, we still recommend using
ValueChoicein favor ofModelParameterChoice, because information are lost when usingModelParameterChoicein exchange of its flexibility, making it incompatible with one-shot strategies and non-python execution engines.Warning
ModelParameterChoicecan NOT be nested.Tip
Although called
ModelParameterChoice, it’s meant to tune hyper-parameter of architecture. It’s NOT used to tune model-training hyper-parameters likelearning_rate. If you need to tunelearning_rate, please useValueChoiceon arguments ofnni.retiarii.Evaluator.- Parameters:
candidates (list of any) – List of values to choose from.
prior (list of float) – Prior distribution to sample from. Currently has no effect.
default (Callable[[List[Any]], Any] or Any) – Function that selects one from
candidates, or a candidate. UseModelParameterChoice.FIRST()orModelParameterChoice.LAST()to take the first or last item. Default:ModelParameterChoice.FIRST()label (str) – Identifier of the value choice.
Warning
ModelParameterChoiceis incompatible with one-shot strategies and non-python execution engines.Sometimes, the same search space implemented without
ModelParameterChoicecan be simpler, and explored with more types of search strategies. For example, the following usages are equivalent:# with ModelParameterChoice depth = nn.ModelParameterChoice(list(range(3, 10))) blocks = [] for i in range(depth): blocks.append(Block()) # w/o HyperParmaeterChoice blocks = Repeat(Block(), (3, 9))
Examples
Get a dynamic-shaped parameter. Because
torch.zerosis not a basic unit, we can’t useValueChoiceon it.>>> parameter_dim = nn.ModelParameterChoice([64, 128, 256]) >>> self.token = nn.Parameter(torch.zeros(1, parameter_dim, 32, 32))
Repeat¶
- class nni.retiarii.nn.pytorch.Repeat(blocks, depth, *, label=None)[source]¶
Repeat a block by a variable number of times.
- Parameters:
blocks (function, list of function, module or list of module) – The block to be repeated. If not a list, it will be replicated (deep-copied) into a list. If a list, it should be of length
max_depth, the modules will be instantiated in order and a prefix will be taken. If a function, it will be called (the argument is the index) to instantiate a module. Otherwise the module will be deep-copied.depth (int or tuple of int) –
If one number, the block will be repeated by a fixed number of times. If a tuple, it should be (min, max), meaning that the block will be repeated at least
mintimes and at mostmaxtimes. If a ValueChoice, it should choose from a series of positive integers.New in version 2.8: Minimum depth can be 0. But this feature is NOT supported on graph engine.
Examples
Block() will be deep copied and repeated 3 times.
self.blocks = nn.Repeat(Block(), 3)
Block() will be repeated 1, 2, or 3 times.
self.blocks = nn.Repeat(Block(), (1, 3))
Can be used together with layer choice. With deep copy, the 3 layers will have the same label, thus share the choice.
self.blocks = nn.Repeat(nn.LayerChoice([...]), (1, 3))
To make the three layer choices independent, we need a factory function that accepts index (0, 1, 2, …) and returns the module of the
index-th layer.self.blocks = nn.Repeat(lambda index: nn.LayerChoice([...], label=f'layer{index}'), (1, 3))
Depth can be a ValueChoice to support arbitrary depth candidate list.
self.blocks = nn.Repeat(Block(), nn.ValueChoice([1, 3, 5]))
Cell¶
- class nni.retiarii.nn.pytorch.Cell(op_candidates, num_nodes, num_ops_per_node=1, num_predecessors=1, merge_op='all', preprocessor=None, postprocessor=None, concat_dim=1, *, label=None)[source]¶
Cell structure that is popularly used in NAS literature.
Find the details in:
On Network Design Spaces for Visual Recognition is a good summary of how this structure works in practice.
A cell consists of multiple “nodes”. Each node is a sum of multiple operators. Each operator is chosen from
op_candidates, and takes one input from previous nodes and predecessors. Predecessor means the input of cell. The output of cell is the concatenation of some of the nodes in the cell (by default all the nodes).Two examples of searched cells are illustrated in the figure below. In these two cells,
op_candidatesare series of convolutions and pooling operations.num_nodes_per_nodeis set to 2.num_nodesis set to 5.merge_opisloose_end. Assuming nodes are enumerated from bottom to top, left to right,output_node_indicesfor the normal cell is[2, 3, 4, 5, 6]. For the reduction cell, it’s[4, 5, 6]. Please take a look at this review article if you are interested in details.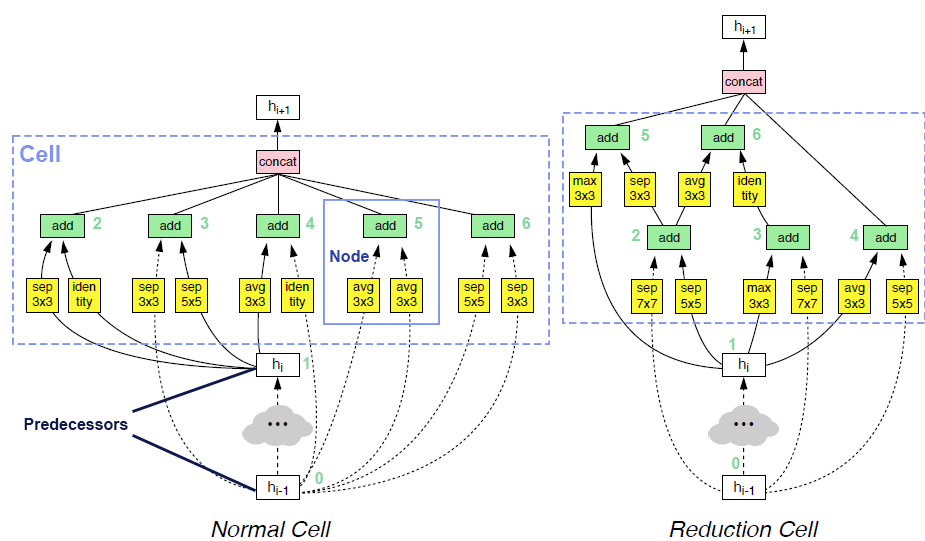
Here is a glossary table, which could help better understand the terms used above:
Name
Brief Description
Cell
A cell consists of
num_nodesnodes.Node
A node is the sum of
num_ops_per_nodeoperators.Operator
Each operator is independently chosen from a list of user-specified candidate operators.
Operator’s input
Each operator has one input, chosen from previous nodes as well as predecessors.
Predecessors
Input of cell. A cell can have multiple predecessors. Predecessors are sent to preprocessor for preprocessing.
Cell’s output
Output of cell. Usually concatenation of some nodes (possibly all nodes) in the cell. Cell’s output, along with predecessors, are sent to postprocessor for postprocessing.
Preprocessor
Extra preprocessing to predecessors. Usually used in shape alignment (e.g., predecessors have different shapes). By default, do nothing.
Postprocessor
Extra postprocessing for cell’s output. Usually used to chain cells with multiple Predecessors (e.g., the next cell wants to have the outputs of both this cell and previous cell as its input). By default, directly use this cell’s output.
Tip
It’s highly recommended to make the candidate operators have an output of the same shape as input. This is because, there can be dynamic connections within cell. If there’s shape change within operations, the input shape of the subsequent operation becomes unknown. In addition, the final concatenation could have shape mismatch issues.
- Parameters:
op_candidates (list of module or function, or dict) – A list of modules to choose from, or a function that accepts current index and optionally its input index, and returns a module. For example, (2, 3, 0) means the 3rd op in the 2nd node, accepts the 0th node as input. The index are enumerated for all nodes including predecessors from 0. When first created, the input index is
None, meaning unknown. Note that in graph execution engine, support of function inop_candidatesis limited. Please also note that, to makeCellwork with one-shot strategy,op_candidates, in case it’s a callable, should not depend on the second input argument, i.e.,op_indexin current node.num_nodes (int) – Number of nodes in the cell.
num_ops_per_node (int) – Number of operators in each node. The output of each node is the sum of all operators in the node. Default: 1.
num_predecessors (int) – Number of inputs of the cell. The input to forward should be a list of tensors. Default: 1.
merge_op ("all", or "loose_end") – If “all”, all the nodes (except predecessors) will be concatenated as the cell’s output, in which case,
output_node_indiceswill belist(range(num_predecessors, num_predecessors + num_nodes)). If “loose_end”, only the nodes that have never been used as other nodes’ inputs will be concatenated to the output. Predecessors are not considered when calculating unused nodes. Details can be found in NDS paper. Default: all.preprocessor (callable) – Override this if some extra transformation on cell’s input is intended. It should be a callable (
nn.Moduleis also acceptable) that takes a list of tensors which are predecessors, and outputs a list of tensors, with the same length as input. By default, it does nothing to the input.postprocessor (callable) – Override this if customization on the output of the cell is intended. It should be a callable that takes the output of this cell, and a list which are predecessors. Its return type should be either one tensor, or a tuple of tensors. The return value of postprocessor is the return value of the cell’s forward. By default, it returns only the output of the current cell.
concat_dim (int) – The result will be a concatenation of several nodes on this dim. Default: 1.
label (str) – Identifier of the cell. Cell sharing the same label will semantically share the same choice.
Examples
Choose between conv2d and maxpool2d. The cell have 4 nodes, 1 op per node, and 2 predecessors.
>>> cell = nn.Cell([nn.Conv2d(32, 32, 3, padding=1), nn.MaxPool2d(3, padding=1)], 4, 1, 2)
In forward:
>>> cell([input1, input2])
The “list bracket” can be omitted:
>>> cell(only_input) # only one input >>> cell(tensor1, tensor2, tensor3) # multiple inputs
Use
merge_opto specify how to construct the output. The output will then have dynamic shape, depending on which input has been used in the cell.>>> cell = nn.Cell([nn.Conv2d(32, 32, 3), nn.MaxPool2d(3)], 4, 1, 2, merge_op='loose_end') >>> cell_out_channels = len(cell.output_node_indices) * 32
The op candidates can be callable that accepts node index in cell, op index in node, and input index.
>>> cell = nn.Cell([ ... lambda node_index, op_index, input_index: nn.Conv2d(32, 32, 3, stride=2 if input_index < 1 else 1), ... ], 4, 1, 2)
Predecessor example:
class Preprocessor: def __init__(self): self.conv1 = nn.Conv2d(16, 32, 1) self.conv2 = nn.Conv2d(64, 32, 1) def forward(self, x): return [self.conv1(x[0]), self.conv2(x[1])] cell = nn.Cell([nn.Conv2d(32, 32, 3), nn.MaxPool2d(3)], 4, 1, 2, preprocessor=Preprocessor()) cell([torch.randn(1, 16, 48, 48), torch.randn(1, 64, 48, 48)]) # the two inputs will be sent to conv1 and conv2 respectively
Warning
Cellis not supported in graph-based execution engine.- output_node_indices¶
An attribute that contains indices of the nodes concatenated to the output (a list of integers).
When the cell is first instantiated in the base model, or when
merge_opisall,output_node_indicesmust berange(num_predecessors, num_predecessors + num_nodes).When
merge_opisloose_end,output_node_indicesis useful to compute the shape of this cell’s output, because the output shape depends on the connection in the cell, and which nodes are “loose ends” depends on mutation.- Type:
list of int
- op_candidates_factory¶
If the operations are created with a factory (callable), this is to be set with the factory. One-shot algorithms will use this to make each node a cartesian product of operations and inputs.
- Type:
CellOpFactory or None
- forward(*inputs)[source]¶
Forward propagation of cell.
- Parameters:
inputs (Union[List[Tensor], Tensor]) – Can be a list of tensors, or several tensors. The length should be equal to
num_predecessors.- Returns:
The return type depends on the output of
postprocessor. By default, it’s the output ofmerge_op, which is a contenation (onconcat_dim) of some of (possibly all) the nodes’ outputs in the cell.- Return type:
Tuple[torch.Tensor] | torch.Tensor
NasBench101Cell¶
- class nni.retiarii.nn.pytorch.NasBench101Cell(op_candidates, in_features, out_features, projection, max_num_nodes=7, max_num_edges=9, label=None)[source]¶
Cell structure that is proposed in NAS-Bench-101.
Proposed by NAS-Bench-101: Towards Reproducible Neural Architecture Search.
This cell is usually used in evaluation of NAS algorithms because there is a “comprehensive analysis” of this search space available, which includes a full architecture-dataset that “maps 423k unique architectures to metrics including run time and accuracy”. You can also use the space in your own space design, in which scenario it should be possible to leverage results in the benchmark to narrow the huge space down to a few efficient architectures.
The space of this cell architecture consists of all possible directed acyclic graphs on no more than
max_num_nodesnodes, where each possible node (other than IN and OUT) has one ofop_candidates, representing the corresponding operation. Edges connecting the nodes can be no more thanmax_num_edges. To align with the paper settings, two vertices specially labeled as operation IN and OUT, are also counted intomax_num_nodesin our implementaion, the default value ofmax_num_nodesis 7 andmax_num_edgesis 9.Input of this cell should be of shape \([N, C_{in}, *]\), while output should be \([N, C_{out}, *]\). The shape of each hidden nodes will be first automatically computed, depending on the cell structure. Each of the
op_candidatesshould be a callable that accepts computednum_featuresand returns aModule. For example,def conv_bn_relu(num_features): return nn.Sequential( nn.Conv2d(num_features, num_features, 1), nn.BatchNorm2d(num_features), nn.ReLU() )
The output of each node is the sum of its input node feed into its operation, except for the last node (output node), which is the concatenation of its input hidden nodes, adding the IN node (if IN and OUT are connected).
When input tensor is added with any other tensor, there could be shape mismatch. Therefore, a projection transformation is needed to transform the input tensor. In paper, this is simply a Conv1x1 followed by BN and ReLU. The
projectionparameters acceptsin_featuresandout_features, returns aModule. This parameter has no default value, as we hold no assumption that users are dealing with images. An example for this parameter is,def projection_fn(in_features, out_features): return nn.Conv2d(in_features, out_features, 1)
- Parameters:
op_candidates (list of callable) – Operation candidates. Each should be a function accepts number of feature, returning nn.Module.
in_features (int) – Input dimension of cell.
out_features (int) – Output dimension of cell.
projection (callable) – Projection module that is used to preprocess the input tensor of the whole cell. A callable that accept input feature and output feature, returning nn.Module.
max_num_nodes (int) – Maximum number of nodes in the cell, input and output included. At least 2. Default: 7.
max_num_edges (int) – Maximum number of edges in the cell. Default: 9.
label (str) – Identifier of the cell. Cell sharing the same label will semantically share the same choice.
Warning
NasBench101Cellis not supported in graph-based execution engine.
NasBench201Cell¶
- class nni.retiarii.nn.pytorch.NasBench201Cell(op_candidates, in_features, out_features, num_tensors=4, label=None)[source]¶
Cell structure that is proposed in NAS-Bench-201.
Proposed by NAS-Bench-201: Extending the Scope of Reproducible Neural Architecture Search.
This cell is a densely connected DAG with
num_tensorsnodes, where each node is tensor. For every i < j, there is an edge from i-th node to j-th node. Each edge in this DAG is associated with an operation transforming the hidden state from the source node to the target node. All possible operations are selected from a predefined operation set, defined inop_candidates. Each of theop_candidatesshould be a callable that accepts input dimension and output dimension, and returns aModule.Input of this cell should be of shape \([N, C_{in}, *]\), while output should be \([N, C_{out}, *]\). For example,
The space size of this cell would be \(|op|^{N(N-1)/2}\), where \(|op|\) is the number of operation candidates, and \(N\) is defined by
num_tensors.- Parameters:
op_candidates (list of callable) – Operation candidates. Each should be a function accepts input feature and output feature, returning nn.Module.
in_features (int) – Input dimension of cell.
out_features (int) – Output dimension of cell.
num_tensors (int) – Number of tensors in the cell (input included). Default: 4
label (str) – Identifier of the cell. Cell sharing the same label will semantically share the same choice.
Hyper-module Library (experimental)¶
AutoActivation¶
- class nni.retiarii.nn.pytorch.AutoActivation(unit_num=1, label=None)[source]¶
This module is an implementation of the paper Searching for Activation Functions.
- Parameters:
unit_num (int) – the number of core units
Notes
Current beta is not per-channel parameter.
Model Space Hub¶
NasBench101¶
- class nni.retiarii.hub.pytorch.NasBench101(*args, **kwargs)[source]¶
The full search space proposed by NAS-Bench-101.
It’s simply a stack of
NasBench101Cell. Operations are conv3x3, conv1x1 and maxpool respectively.- Parameters:
stem_out_channels (int) – Number of output channels of the stem convolution.
num_stacks (int) – Number of stacks in the network.
num_modules_per_stack (int) – Number of modules in each stack. Each module is a
NasBench101Cell.max_num_vertices (int) – Maximum number of vertices in each cell.
max_num_edges (int) – Maximum number of edges in each cell.
num_labels (int) – Number of categories for classification.
bn_eps (float) – Epsilon for batch normalization.
bn_momentum (float) – Momentum for batch normalization.
NasBench201¶
- class nni.retiarii.hub.pytorch.NasBench201(*args, **kwargs)[source]¶
The full search space proposed by NAS-Bench-201.
It’s a stack of
NasBench201Cell.- Parameters:
stem_out_channels (int) – The output channels of the stem.
num_modules_per_stack (int) – The number of modules (cells) in each stack. Each cell is a
NasBench201Cell.num_labels (int) – Number of categories for classification.
NASNet¶
- class nni.retiarii.hub.pytorch.NASNet(*args, **kwargs)[source]¶
Search space proposed in Learning Transferable Architectures for Scalable Image Recognition.
It is built upon
Cell, and implemented based onNDS. Its operator candidates areNASNET_OPS. It has 5 nodes per cell, and the output is concatenation of nodes not used as input to other nodes.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.retiarii.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width (Union[Tuple[int, ...], int]) – A fixed initial width or a tuple of widths to choose from.
num_cells (Union[Tuple[int, ...], int]) – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset (Literal['cifar', 'imagenet']) – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss (bool) – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob (float) – Apply drop path. Enabled when it’s set to be greater than 0.
- class nni.retiarii.hub.pytorch.nasnet.NDS(op_candidates, merge_op='all', num_nodes_per_cell=4, width=16, num_cells=20, dataset='imagenet', auxiliary_loss=False, drop_path_prob=0.0)[source]¶
The unified version of NASNet search space.
We follow the implementation in unnas. See On Network Design Spaces for Visual Recognition for details.
Different NAS papers usually differ in the way that they specify
op_candidatesandmerge_op.datasethere is to give a hint about input resolution, so as to create reasonable stem and auxiliary heads.NDS has a speciality that it has mutable depths/widths. This is implemented by accepting a list of int as
num_cells/width.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.retiarii.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width (Union[Tuple[int, ...], int]) – A fixed initial width or a tuple of widths to choose from.
num_cells (Union[Tuple[int, ...], int]) – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset (Literal['cifar', 'imagenet']) – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss (bool) – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob (float) – Apply drop path. Enabled when it’s set to be greater than 0.
op_candidates (List[str]) – List of operator candidates. Must be from
OPS.merge_op (Literal['all', 'loose_end']) – See
Cell.num_nodes_per_cell (int) – See
Cell.
- set_drop_path_prob(drop_prob)[source]¶
Set the drop probability of Drop-path in the network. Reference: FractalNet: Ultra-Deep Neural Networks without Residuals.
- class nni.retiarii.hub.pytorch.nasnet.NDSStage(blocks, depth, *, label=None)[source]¶
This class defines NDSStage, a special type of Repeat, for isinstance check, and shape alignment.
In NDS, we can’t simply use Repeat to stack the blocks, because the output shape of each stacked block can be different. This is a problem for one-shot strategy because they assume every possible candidate should return values of the same shape.
Therefore, we need
NDSStagePathSamplingandNDSStageDifferentiableto manually align the shapes – specifically, to transform the first block in each stage.This is not required though, when depth is not changing, or the mutable depth causes no problem (e.g., when the minimum depth is large enough).
Attention
Assumption: Loose end is treated as all in
merge_op(the case in one-shot), which enforces reduction cell and normal cells in the same stage to have the exact same output shape.- downsampling: bool¶
This stage has downsampling
- estimated_out_channels: int¶
Output channels of this stage. It’s estimated because it assumes
allasmerge_op.
- estimated_out_channels_prev: int¶
Output channels of cells in last stage.
ENAS¶
- class nni.retiarii.hub.pytorch.ENAS(*args, **kwargs)[source]¶
Search space proposed in Efficient neural architecture search via parameter sharing.
It is built upon
Cell, and implemented based onNDS. Its operator candidates areENAS_OPS. It has 5 nodes per cell, and the output is concatenation of nodes not used as input to other nodes.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.retiarii.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width (Union[Tuple[int, ...], int]) – A fixed initial width or a tuple of widths to choose from.
num_cells (Union[Tuple[int, ...], int]) – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset (Literal['cifar', 'imagenet']) – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss (bool) – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob (float) – Apply drop path. Enabled when it’s set to be greater than 0.
AmoebaNet¶
- class nni.retiarii.hub.pytorch.AmoebaNet(*args, **kwargs)[source]¶
Search space proposed in Regularized evolution for image classifier architecture search.
It is built upon
Cell, and implemented based onNDS. Its operator candidates areAMOEBA_OPS. It has 5 nodes per cell, and the output is concatenation of nodes not used as input to other nodes.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.retiarii.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width (Union[Tuple[int, ...], int]) – A fixed initial width or a tuple of widths to choose from.
num_cells (Union[Tuple[int, ...], int]) – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset (Literal['cifar', 'imagenet']) – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss (bool) – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob (float) – Apply drop path. Enabled when it’s set to be greater than 0.
PNAS¶
- class nni.retiarii.hub.pytorch.PNAS(*args, **kwargs)[source]¶
Search space proposed in Progressive neural architecture search.
It is built upon
Cell, and implemented based onNDS. Its operator candidates arePNAS_OPS. It has 5 nodes per cell, and the output is concatenation of all nodes in the cell.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.retiarii.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width (Union[Tuple[int, ...], int]) – A fixed initial width or a tuple of widths to choose from.
num_cells (Union[Tuple[int, ...], int]) – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset (Literal['cifar', 'imagenet']) – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss (bool) – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob (float) – Apply drop path. Enabled when it’s set to be greater than 0.
DARTS¶
- class nni.retiarii.hub.pytorch.DARTS(*args, **kwargs)[source]¶
Search space proposed in Darts: Differentiable architecture search.
It is built upon
Cell, and implemented based onNDS. Its operator candidates areDARTS_OPS. It has 4 nodes per cell, and the output is concatenation of all nodes in the cell.Note
noneis not included in the operator candidates. It has already been handled in the differentiable implementation of cell.Notes
To use NDS spaces with one-shot strategies, especially when depth is mutating (i.e.,
num_cellsis set to a tuple / list), please useNDSStagePathSampling(with ENAS and RandomOneShot) andNDSStageDifferentiable(with DARTS and Proxyless) intomutation_hooks. This is because the output shape of each stacked block inNDSStagecan be different. For example:from nni.retiarii.hub.pytorch.nasnet import NDSStageDifferentiable darts_strategy = strategy.DARTS(mutation_hooks=[NDSStageDifferentiable.mutate])
- Parameters:
width (Union[Tuple[int, ...], int]) – A fixed initial width or a tuple of widths to choose from.
num_cells (Union[Tuple[int, ...], int]) – A fixed number of cells (depths) to stack, or a tuple of depths to choose from.
dataset (Literal['cifar', 'imagenet']) – The essential differences are in “stem” cells, i.e., how they process the raw image input. Choosing “imagenet” means more downsampling at the beginning of the network.
auxiliary_loss (bool) – If true, another auxiliary classification head will produce the another prediction. This makes the output of network two logits in the training phase.
drop_path_prob (float) – Apply drop path. Enabled when it’s set to be greater than 0.
ProxylessNAS¶
- class nni.retiarii.hub.pytorch.ProxylessNAS(*args, **kwargs)[source]¶
The search space proposed by ProxylessNAS.
Following the official implementation, the inverted residual with kernel size / expand ratio variations in each layer is implemented with a
LayerChoicewith all-combination candidates. That means, when used in weight sharing, these candidates will be treated as separate layers, and won’t be fine-grained shared. We note thatMobileNetV3Spaceis different in this perspective.This space can be implemented as part of
MobileNetV3Space, but we separate those following conventions.- Parameters:
num_labels (int) – The number of labels for classification.
base_widths (Tuple[int, ...]) – Widths of each stage, from stem, to body, to head. Length should be 9.
dropout_rate (float) – Dropout rate for the final classification layer.
width_mult (float) – Width multiplier for the model.
bn_eps (float) – Epsilon for batch normalization.
bn_momentum (float) – Momentum for batch normalization.
- class nni.retiarii.hub.pytorch.proxylessnas.InvertedResidual(in_channels, out_channels, expand_ratio, kernel_size=3, stride=1, squeeze_excite=None, norm_layer=None, activation_layer=None)[source]¶
An Inverted Residual Block, sometimes called an MBConv Block, is a type of residual block used for image models that uses an inverted structure for efficiency reasons.
It was originally proposed for the MobileNetV2 CNN architecture. It has since been reused for several mobile-optimized CNNs. It follows a narrow -> wide -> narrow approach, hence the inversion. It first widens with a 1x1 convolution, then uses a 3x3 depthwise convolution (which greatly reduces the number of parameters), then a 1x1 convolution is used to reduce the number of channels so input and output can be added.
This implementation is sort of a mixture between:
- Parameters:
in_channels (Union[ValueChoiceX[int], int]) – The number of input channels. Can be a value choice.
out_channels (Union[ValueChoiceX[int], int]) – The number of output channels. Can be a value choice.
expand_ratio (Union[ValueChoiceX[float], float]) – The ratio of intermediate channels with respect to input channels. Can be a value choice.
kernel_size (Union[ValueChoiceX[int], int]) – The kernel size of the depthwise convolution. Can be a value choice.
stride (int) – The stride of the depthwise convolution.
squeeze_excite (Optional[Callable[[Union[ValueChoiceX[int], int], Union[ValueChoiceX[int], int]], Module]]) – Callable to create squeeze and excitation layer. Take hidden channels and input channels as arguments.
norm_layer (Optional[Callable[[int], Module]]) – Callable to create normalization layer. Take input channels as argument.
activation_layer (Optional[Callable[[...], Module]]) – Callable to create activation layer. No input arguments.
MobileNetV3Space¶
- class nni.retiarii.hub.pytorch.MobileNetV3Space(*args, **kwargs)[source]¶
MobileNetV3Space implements the largest search space in TuNAS.
The search dimensions include widths, expand ratios, kernel sizes, SE ratio. Some of them can be turned off via arguments to narrow down the search space.
Different from ProxylessNAS search space, this space is implemented with
ValueChoice.We use the following snipppet as reference. https://github.com/google-research/google-research/blob/20736344591f774f4b1570af64624ed1e18d2867/tunas/mobile_search_space_v3.py#L728
We have
num_blockswhich equals to the length ofself.blocks(the main body of the network). For simplicity, the following parameter specification assumesnum_blocksequals 8 (body + head). If a shallower body is intended, arrays includingbase_widths,squeeze_excite,depth_range,stride,activationshould also be shortened accordingly.- Parameters:
num_labels (int) – Dimensions for classification head.
base_widths (Tuple[int, ...]) – Widths of each stage, from stem, to body, to head. Length should be 9, i.e.,
num_blocks + 1(because there is a stem width in front).width_multipliers (Union[Tuple[float, ...], float]) – A range of widths multiplier to choose from. The choice is independent for each stage. Or it can be a fixed float. This will be applied on
base_widths, and we would also make sure that widths can be divided by 8.expand_ratios (Tuple[float, ...]) – A list of expand ratios to choose from. Independent for every block.
squeeze_excite (Tuple[Literal['force', 'optional', 'none'], ...]) – Indicating whether the current stage can have an optional SE layer. Expect array of length 6 for stage 0 to 5. Each element can be one of
force,optional,none.depth_range (List[Tuple[int, int]]) – A range (e.g.,
(1, 4)), or a list of range (e.g.,[(1, 3), (1, 4), (1, 4), (1, 3), (0, 2)]). If a list, the length should be 5. The depth are specified for stage 1 to 5.stride (Tuple[int, ...]) – Stride for all stages (including stem and head). Length should be same as
base_widths.activation (Tuple[Literal['hswish', 'swish', 'relu'], ...]) – Activation (class) for all stages. Length is same as
base_widths.se_from_exp (bool) – Calculate SE channel reduction from expanded (mid) channels.
dropout_rate (float) – Dropout rate at classification head.
bn_eps (float) – Epsilon of batch normalization.
bn_momentum (float) – Momentum of batch normalization.
ShuffleNetSpace¶
- class nni.retiarii.hub.pytorch.ShuffleNetSpace(*args, **kwargs)[source]¶
The search space proposed in Single Path One-shot.
The basic building block design is inspired by a state-of-the-art manually-designed network – ShuffleNetV2. There are 20 choice blocks in total. Each choice block has 4 candidates, namely
choice 3,choice 5,choice_7andchoice_xrespectively. They differ in kernel sizes and the number of depthwise convolutions. The size of the search space is \(4^{20}\).- Parameters:
num_labels (int) – Number of classes for the classification head. Default: 1000.
channel_search (bool) – If true, for each building block, the number of
mid_channels(output channels of the first 1x1 conv in each building block) varies from 0.2x to 1.6x (quantized to multiple of 0.2). Here, “k-x” means k times the number of default channels. Otherwise, 1.0x is used by default. Default: false.affine (bool) – Apply affine to all batch norm. Default: true.
AutoformerSpace¶
- class nni.retiarii.hub.pytorch.AutoformerSpace(*args, **kwargs)[source]¶
The search space that is proposed in Autoformer. There are four searchable variables: depth, embedding dimension, heads number and MLP ratio.
- Parameters:
search_embed_dim (list of int) – The search space of embedding dimension.
search_mlp_ratio (list of float) – The search space of MLP ratio.
search_num_heads (list of int) – The search space of number of heads.
search_depth (list of int) – The search space of depth.
img_size (int) – Size of input image.
patch_size (int) – Size of image patch.
in_chans (int) – Number of channels of the input image.
num_classes (int) – Number of classes for classifier.
qkv_bias (bool) – Whether to use bias item in the qkv embedding.
drop_rate (float) – Drop rate of the MLP projection in MSA and FFN.
attn_drop_rate (float) – Drop rate of attention.
drop_path_rate (float) – Drop path rate.
pre_norm (bool) – Whether to use pre_norm. Otherwise post_norm is used.
global_pool (bool) – Whether to use global pooling to generate the image representation. Otherwise the cls_token is used.
abs_pos (bool) – Whether to use absolute positional embeddings.
qk_scale (float) – The scaler on score map in self-attention.
rpe (bool) – Whether to use relative position encoding.
Mutators (advanced)¶
Mutator¶
- class nni.retiarii.Mutator(sampler=None, label=None)[source]¶
Mutates graphs in model to generate new model. Mutator class will be used in two places:
Inherit Mutator to implement graph mutation logic.
Use Mutator subclass to implement NAS strategy.
In scenario 1, the subclass should implement Mutator.mutate() interface with Mutator.choice(). In scenario 2, strategy should use constructor or Mutator.bind_sampler() to initialize subclass, and then use Mutator.apply() to mutate model. For certain mutator subclasses, strategy or sampler can use Mutator.dry_run() to predict choice candidates. # Method names are open for discussion.
If mutator has a label, in most cases, it means that this mutator is applied to nodes with this label.
- apply(model)[source]¶
Apply this mutator on a model. Returns mutated model. The model will be copied before mutation and the original model will not be modified.
Placeholder¶
Graph¶
- class nni.retiarii.Model(_internal=False)[source]¶
Represents a neural network model.
During mutation, one
Modelobject is created for each trainable snapshot. For example, consider a mutator that insert a node at an edge for each iteration. In one iteration, the mutator invokes 4 primitives: add node, remove edge, add edge to head, add edge to tail. These 4 primitives operates in oneModelobject. When they are all done the model will be set to “frozen” (trainable) status and be submitted to execution engine. And then a new iteration starts, and a newModelobject is created by forking last model.- python_object¶
Python object of base model. It will be none when the base model is not available.
- python_class¶
Python class that base model is converted from.
- python_init_params¶
Initialization parameters of python class.
- status¶
See
ModelStatus.
- root_graph¶
The outermost graph which usually takes dataset as input and feeds output to loss function.
- graphs¶
All graphs (subgraphs) in this model.
- evaluator¶
Model evaluator
- history¶
Mutation history.
selfis directly mutated fromself.history[-1];self.history[-1]is mutated fromself.history[-2], and so on.self.history[0]is the base graph.
- metric¶
Training result of the model, or
Noneif it’s not yet trained or has failed to train.
- intermediate_metrics¶
Intermediate training metrics. If the model is not trained, it’s an empty list.
- fork()[source]¶
Create a new model which has same topology, names, and IDs to current one.
Can only be invoked on a frozen model. The new model will be in Mutating state.
This API is used in mutator base class.
- get_node_by_name(node_name)[source]¶
Traverse all the nodes to find the matched node with the given name.
- get_node_by_python_name(python_name)[source]¶
Traverse all the nodes to find the matched node with the given python_name.
- class nni.retiarii.Graph(model, graph_id, name=None, _internal=False)[source]¶
Graph topology.
This class simply represents the topology, with no semantic meaning. All other information like metric, non-graph functions, mutation history, etc should go to
Model.Each graph belongs to and only belongs to one
Model.- model¶
The model containing (and owning) this graph.
- id¶
Unique ID in the model. If two models have graphs of identical ID, they are semantically the same graph. Typically this means one graph is mutated from another, or they are both mutated from one ancestor.
- name¶
Mnemonic name of this graph. It should have an one-to-one mapping with ID.
- input_names¶
Optional mnemonic names of input parameters.
- output_names¶
Optional mnemonic names of output values.
- input_node¶
Incoming node.
- output_node¶
Output node.
Hidden nodes
- nodes¶
All input/output/hidden nodes.
- edges¶
Edges.
- python_name¶
The name of torch.nn.Module, should have one-to-one mapping with items in python model.
- fork()[source]¶
Fork the model and returns corresponding graph in new model. This shortcut might be helpful because many algorithms only cares about “stem” subgraph instead of whole model.
- get_node_by_id(node_id)[source]¶
Returns the node which has specified name; or returns None if no node has this name.
- get_node_by_name(name)[source]¶
Returns the node which has specified name; or returns None if no node has this name.
- class nni.retiarii.Node(graph, node_id, name, operation, _internal=False)[source]¶
An operation or an opaque subgraph inside a graph.
Each node belongs to and only belongs to one
Graph. Nodes should never be created with constructor. UseGraph.add_node()instead.The node itself is for topology only. Information of tensor calculation should all go inside
operationattribute.TODO: parameter of subgraph (cell) It’s easy to assign parameters on cell node, but it’s hard to “use” them. We need to design a way to reference stored cell parameters in inner node operations. e.g.
self.fc = Linear(self.units)<- how to expressself.unitsin IR?- graph¶
The graph containing this node.
- id¶
Unique ID in the model. If two models have nodes with same ID, they are semantically the same node.
- name¶
Mnemonic name. It should have an one-to-one mapping with ID.
- python_name¶
The name of torch.nn.Module, should have one-to-one mapping with items in python model.
- label¶
Optional. If two nodes have the same label, they are considered same by the mutator.
- operation¶
Operation.
- cell¶
Read only shortcut to get the referenced subgraph. If this node is not a subgraph (is a primitive operation), accessing
cellwill raise an error.
- predecessors¶
Predecessor nodes of this node in the graph. This is an optional mutation helper.
- successors¶
Successor nodes of this node in the graph. This is an optional mutation helper.
- incoming_edges¶
Incoming edges of this node in the graph. This is an optional mutation helper.
- outgoing_edges¶
Outgoing edges of this node in the graph. This is an optional mutation helper.
- class nni.retiarii.Edge(head, tail, _internal=False)[source]¶
A tensor, or “data flow”, between two nodes.
Example forward code snippet:
a, b, c = split(x) p = concat(a, c) q = sum(b, p) z = relu(q)
Edges in above snippet:
+ head: (split, 0), tail: (concat, 0) # a in concat + head: (split, 2), tail: (concat, 1) # c in concat + head: (split, 1), tail: (sum, -1 or 0) # b in sum + head: (concat, null), tail: (sum, -1 or 1) # p in sum + head: (sum, null), tail: (relu, null) # q in relu
- graph¶
Graph.
- head¶
Head node.
- tail¶
Tail node.
- head_slot¶
Index of outputs in head node. If the node has only one output, this should be
null.
- tail_slot¶
Index of inputs in tail node. If the node has only one input, this should be
null. If the node does not care about order, this can be-1.
- class nni.retiarii.Operation(type_name, parameters={}, _internal=False, attributes={})[source]¶
Calculation logic of a graph node.
The constructor is private. Use Operation.new() to create operation object.
Operation is a naive record. Do not “mutate” its attributes or store information relate to specific node. All complex logic should be implemented in Node class.
- type¶
Operation type name (e.g. Conv2D). If it starts with underscore, the “operation” is a special one (e.g. subgraph, input/output).
- parameters¶
Arbitrary key-value parameters (e.g. kernel_size).