Note
Click here to download the full example code
Hello, NAS!¶
This is the 101 tutorial of Neural Architecture Search (NAS) on NNI. In this tutorial, we will search for a neural architecture on MNIST dataset with the help of NAS framework of NNI, i.e., Retiarii. We use multi-trial NAS as an example to show how to construct and explore a model space.
There are mainly three crucial components for a neural architecture search task, namely,
Model search space that defines a set of models to explore.
A proper strategy as the method to explore this model space.
A model evaluator that reports the performance of every model in the space.
Currently, PyTorch is the only supported framework by Retiarii, and we have only tested PyTorch 1.7 to 1.10. This tutorial assumes PyTorch context but it should also apply to other frameworks, which is in our future plan.
Define your Model Space¶
Model space is defined by users to express a set of models that users want to explore, which contains potentially good-performing models. In this framework, a model space is defined with two parts: a base model and possible mutations on the base model.
Define Base Model¶
Defining a base model is almost the same as defining a PyTorch (or TensorFlow) model.
Usually, you only need to replace the code import torch.nn as nn with
import nni.retiarii.nn.pytorch as nn to use our wrapped PyTorch modules.
Below is a very simple example of defining a base model.
import torch
import torch.nn.functional as F
import nni.retiarii.nn.pytorch as nn
from nni.retiarii import model_wrapper
@model_wrapper # this decorator should be put on the out most
class Net(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1, 32, 3, 1)
self.conv2 = nn.Conv2d(32, 64, 3, 1)
self.dropout1 = nn.Dropout(0.25)
self.dropout2 = nn.Dropout(0.5)
self.fc1 = nn.Linear(9216, 128)
self.fc2 = nn.Linear(128, 10)
def forward(self, x):
x = F.relu(self.conv1(x))
x = F.max_pool2d(self.conv2(x), 2)
x = torch.flatten(self.dropout1(x), 1)
x = self.fc2(self.dropout2(F.relu(self.fc1(x))))
output = F.log_softmax(x, dim=1)
return output
Tip
Always keep in mind that you should use import nni.retiarii.nn.pytorch as nn and nni.retiarii.model_wrapper().
Many mistakes are a result of forgetting one of those.
Also, please use torch.nn for submodules of nn.init, e.g., torch.nn.init instead of nn.init.
Define Model Mutations¶
A base model is only one concrete model not a model space. We provide API and Primitives for users to express how the base model can be mutated. That is, to build a model space which includes many models.
Based on the above base model, we can define a model space as below.
@model_wrapper
class Net(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1, 32, 3, 1)
- self.conv2 = nn.Conv2d(32, 64, 3, 1)
+ self.conv2 = nn.LayerChoice([
+ nn.Conv2d(32, 64, 3, 1),
+ DepthwiseSeparableConv(32, 64)
+ ])
- self.dropout1 = nn.Dropout(0.25)
+ self.dropout1 = nn.Dropout(nn.ValueChoice([0.25, 0.5, 0.75]))
self.dropout2 = nn.Dropout(0.5)
- self.fc1 = nn.Linear(9216, 128)
- self.fc2 = nn.Linear(128, 10)
+ feature = nn.ValueChoice([64, 128, 256])
+ self.fc1 = nn.Linear(9216, feature)
+ self.fc2 = nn.Linear(feature, 10)
def forward(self, x):
x = F.relu(self.conv1(x))
x = F.max_pool2d(self.conv2(x), 2)
x = torch.flatten(self.dropout1(x), 1)
x = self.fc2(self.dropout2(F.relu(self.fc1(x))))
output = F.log_softmax(x, dim=1)
return output
This results in the following code:
class DepthwiseSeparableConv(nn.Module):
def __init__(self, in_ch, out_ch):
super().__init__()
self.depthwise = nn.Conv2d(in_ch, in_ch, kernel_size=3, groups=in_ch)
self.pointwise = nn.Conv2d(in_ch, out_ch, kernel_size=1)
def forward(self, x):
return self.pointwise(self.depthwise(x))
@model_wrapper
class ModelSpace(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1, 32, 3, 1)
# LayerChoice is used to select a layer between Conv2d and DwConv.
self.conv2 = nn.LayerChoice([
nn.Conv2d(32, 64, 3, 1),
DepthwiseSeparableConv(32, 64)
])
# ValueChoice is used to select a dropout rate.
# ValueChoice can be used as parameter of modules wrapped in `nni.retiarii.nn.pytorch`
# or customized modules wrapped with `@basic_unit`.
self.dropout1 = nn.Dropout(nn.ValueChoice([0.25, 0.5, 0.75])) # choose dropout rate from 0.25, 0.5 and 0.75
self.dropout2 = nn.Dropout(0.5)
feature = nn.ValueChoice([64, 128, 256])
self.fc1 = nn.Linear(9216, feature)
self.fc2 = nn.Linear(feature, 10)
def forward(self, x):
x = F.relu(self.conv1(x))
x = F.max_pool2d(self.conv2(x), 2)
x = torch.flatten(self.dropout1(x), 1)
x = self.fc2(self.dropout2(F.relu(self.fc1(x))))
output = F.log_softmax(x, dim=1)
return output
model_space = ModelSpace()
model_space
Out:
ModelSpace(
(conv1): Conv2d(1, 32, kernel_size=(3, 3), stride=(1, 1))
(conv2): LayerChoice([Conv2d(32, 64, kernel_size=(3, 3), stride=(1, 1)), DepthwiseSeparableConv(
(depthwise): Conv2d(32, 32, kernel_size=(3, 3), stride=(1, 1), groups=32)
(pointwise): Conv2d(32, 64, kernel_size=(1, 1), stride=(1, 1))
)], label='model_1')
(dropout1): Dropout(p=0.25, inplace=False)
(dropout2): Dropout(p=0.5, inplace=False)
(fc1): Linear(in_features=9216, out_features=64, bias=True)
(fc2): Linear(in_features=64, out_features=10, bias=True)
)
This example uses two mutation APIs,
nn.LayerChoice and
nn.InputChoice.
nn.LayerChoice
takes a list of candidate modules (two in this example), one will be chosen for each sampled model.
It can be used like normal PyTorch module.
nn.InputChoice takes a list of candidate values,
one will be chosen to take effect for each sampled model.
More detailed API description and usage can be found here.
Note
We are actively enriching the mutation APIs, to facilitate easy construction of model space. If the currently supported mutation APIs cannot express your model space, please refer to this doc for customizing mutators.
Explore the Defined Model Space¶
There are basically two exploration approaches: (1) search by evaluating each sampled model independently, which is the search approach in multi-trial NAS and (2) one-shot weight-sharing based search, which is used in one-shot NAS. We demonstrate the first approach in this tutorial. Users can refer to here for the second approach.
First, users need to pick a proper exploration strategy to explore the defined model space. Second, users need to pick or customize a model evaluator to evaluate the performance of each explored model.
Pick an exploration strategy¶
Retiarii supports many exploration strategies.
Simply choosing (i.e., instantiate) an exploration strategy as below.
import nni.retiarii.strategy as strategy
search_strategy = strategy.Random(dedup=True) # dedup=False if deduplication is not wanted
Out:
/home/yugzhan/miniconda3/envs/cu102/lib/python3.8/site-packages/ray/autoscaler/_private/cli_logger.py:57: FutureWarning: Not all Ray CLI dependencies were found. In Ray 1.4+, the Ray CLI, autoscaler, and dashboard will only be usable via `pip install 'ray[default]'`. Please update your install command.
warnings.warn(
Pick or customize a model evaluator¶
In the exploration process, the exploration strategy repeatedly generates new models. A model evaluator is for training and validating each generated model to obtain the model’s performance. The performance is sent to the exploration strategy for the strategy to generate better models.
Retiarii has provided built-in model evaluators, but to start with,
it is recommended to use FunctionalEvaluator,
that is, to wrap your own training and evaluation code with one single function.
This function should receive one single model class and uses nni.report_final_result() to report the final score of this model.
An example here creates a simple evaluator that runs on MNIST dataset, trains for 2 epochs, and reports its validation accuracy.
import nni
from torchvision import transforms
from torchvision.datasets import MNIST
from torch.utils.data import DataLoader
def train_epoch(model, device, train_loader, optimizer, epoch):
loss_fn = torch.nn.CrossEntropyLoss()
model.train()
for batch_idx, (data, target) in enumerate(train_loader):
data, target = data.to(device), target.to(device)
optimizer.zero_grad()
output = model(data)
loss = loss_fn(output, target)
loss.backward()
optimizer.step()
if batch_idx % 10 == 0:
print('Train Epoch: {} [{}/{} ({:.0f}%)]\tLoss: {:.6f}'.format(
epoch, batch_idx * len(data), len(train_loader.dataset),
100. * batch_idx / len(train_loader), loss.item()))
def test_epoch(model, device, test_loader):
model.eval()
test_loss = 0
correct = 0
with torch.no_grad():
for data, target in test_loader:
data, target = data.to(device), target.to(device)
output = model(data)
pred = output.argmax(dim=1, keepdim=True)
correct += pred.eq(target.view_as(pred)).sum().item()
test_loss /= len(test_loader.dataset)
accuracy = 100. * correct / len(test_loader.dataset)
print('\nTest set: Accuracy: {}/{} ({:.0f}%)\n'.format(
correct, len(test_loader.dataset), accuracy))
return accuracy
def evaluate_model(model_cls):
# "model_cls" is a class, need to instantiate
model = model_cls()
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
model.to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3)
transf = transforms.Compose([transforms.ToTensor(), transforms.Normalize((0.1307,), (0.3081,))])
train_loader = DataLoader(MNIST('data/mnist', download=True, transform=transf), batch_size=64, shuffle=True)
test_loader = DataLoader(MNIST('data/mnist', download=True, train=False, transform=transf), batch_size=64)
for epoch in range(3):
# train the model for one epoch
train_epoch(model, device, train_loader, optimizer, epoch)
# test the model for one epoch
accuracy = test_epoch(model, device, test_loader)
# call report intermediate result. Result can be float or dict
nni.report_intermediate_result(accuracy)
# report final test result
nni.report_final_result(accuracy)
Create the evaluator
from nni.retiarii.evaluator import FunctionalEvaluator
evaluator = FunctionalEvaluator(evaluate_model)
The train_epoch and test_epoch here can be any customized function,
where users can write their own training recipe.
It is recommended that the evaluate_model here accepts no additional arguments other than model_cls.
However, in the advanced tutorial, we will show how to use additional arguments in case you actually need those.
In future, we will support mutation on the arguments of evaluators, which is commonly called “Hyper-parmeter tuning”.
Launch an Experiment¶
After all the above are prepared, it is time to start an experiment to do the model search. An example is shown below.
from nni.retiarii.experiment.pytorch import RetiariiExperiment, RetiariiExeConfig
exp = RetiariiExperiment(model_space, evaluator, [], search_strategy)
exp_config = RetiariiExeConfig('local')
exp_config.experiment_name = 'mnist_search'
The following configurations are useful to control how many trials to run at most / at the same time.
exp_config.max_trial_number = 4 # spawn 4 trials at most
exp_config.trial_concurrency = 2 # will run two trials concurrently
Remember to set the following config if you want to GPU.
use_active_gpu should be set true if you wish to use an occupied GPU (possibly running a GUI).
exp_config.trial_gpu_number = 1
exp_config.training_service.use_active_gpu = True
Launch the experiment. The experiment should take several minutes to finish on a workstation with 2 GPUs.
exp.run(exp_config, 8081)
Out:
INFO:nni.experiment:Creating experiment, Experiment ID: z8ns5fv7
INFO:nni.experiment:Connecting IPC pipe...
INFO:nni.experiment:Starting web server...
INFO:nni.experiment:Setting up...
INFO:nni.runtime.msg_dispatcher_base:Dispatcher started
INFO:nni.retiarii.experiment.pytorch:Web UI URLs: http://127.0.0.1:8081 http://10.190.172.35:8081 http://192.168.49.1:8081 http://172.17.0.1:8081
INFO:nni.retiarii.experiment.pytorch:Start strategy...
INFO:root:Successfully update searchSpace.
INFO:nni.retiarii.strategy.bruteforce:Random search running in fixed size mode. Dedup: on.
INFO:nni.retiarii.experiment.pytorch:Stopping experiment, please wait...
INFO:nni.retiarii.experiment.pytorch:Strategy exit
INFO:nni.retiarii.experiment.pytorch:Waiting for experiment to become DONE (you can ctrl+c if there is no running trial jobs)...
INFO:nni.runtime.msg_dispatcher_base:Dispatcher exiting...
INFO:nni.retiarii.experiment.pytorch:Experiment stopped
Users can also run Retiarii Experiment with different training services
besides local training service.
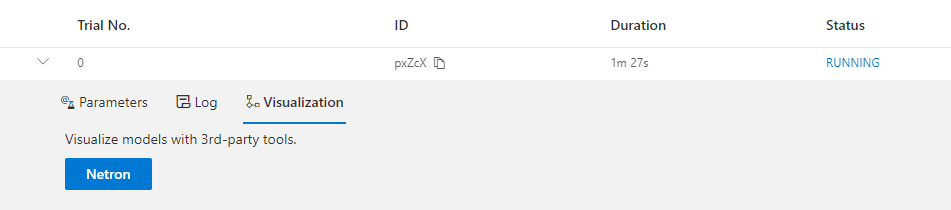
Visualize the Experiment¶
Users can visualize their experiment in the same way as visualizing a normal hyper-parameter tuning experiment.
For example, open localhost:8081 in your browser, 8081 is the port that you set in exp.run.
Please refer to here for details.
We support visualizing models with 3rd-party visualization engines (like Netron).
This can be used by clicking Visualization in detail panel for each trial.
Note that current visualization is based on onnx ,
thus visualization is not feasible if the model cannot be exported into onnx.
Built-in evaluators (e.g., Classification) will automatically export the model into a file.
For your own evaluator, you need to save your file into $NNI_OUTPUT_DIR/model.onnx to make this work.
For instance,
import os
from pathlib import Path
def evaluate_model_with_visualization(model_cls):
model = model_cls()
# dump the model into an onnx
if 'NNI_OUTPUT_DIR' in os.environ:
dummy_input = torch.zeros(1, 3, 32, 32)
torch.onnx.export(model, (dummy_input, ),
Path(os.environ['NNI_OUTPUT_DIR']) / 'model.onnx')
evaluate_model(model_cls)
Relaunch the experiment, and a button is shown on Web portal.
Export Top Models¶
Users can export top models after the exploration is done using export_top_models.
for model_dict in exp.export_top_models(formatter='dict'):
print(model_dict)
Out:
{'model_1': '0', 'model_2': 0.25, 'model_3': 64}
The output is json object which records the mutation actions of the top model.
If users want to output source code of the top model,
they can use graph-based execution engine for the experiment,
by simply adding the following two lines.
exp_config.execution_engine = 'base'
export_formatter = 'code'
Total running time of the script: ( 2 minutes 4.499 seconds)